Uni-Mol2 Release: Joining Hands with the DeepModeling Community, Marching Towards a 3D Molecular Foundation Model
DeepModeling community has officially released Uni-Mol2, which is currently the largest 3D molecular representation foundation model. The largest version of Uni-Mol2 has a parameter scale of 1.1 billion and has been pre-trained on 800 million molecular conformations, demonstrating excellent performance in multiple molecular property prediction tasks. This achievement not only provides a powerful tool for deep learning research in the field of molecular science but also lays a solid experimental foundation for exploring larger-scale molecular pre-training models. At the NeurIPS 2024 conference currently being held in Vancouver, Canada, Uni-Mol2, as an accepted paper, has also received extensive attention.
Version Release Content
Better Prediction Performance
The research team has proposed an innovative "dual-track Transformer" architecture. By separately processing atomic-level, graph-level, and molecular conformation-level features, multi-dimensional modeling of molecular information is achieved. The core of this architecture lies in simultaneously processing atomic-level features and atomic-pair-level features, thus achieving a comprehensive representation of molecular information. Specifically, the backbone network of Uni-Mol2 updates atomic representations and atomic-pair representations in parallel in each module and realizes the deep fusion of features through an attention mechanism with atomic-pair bias. This design provides a powerful expressive ability for modeling complex molecular structures.
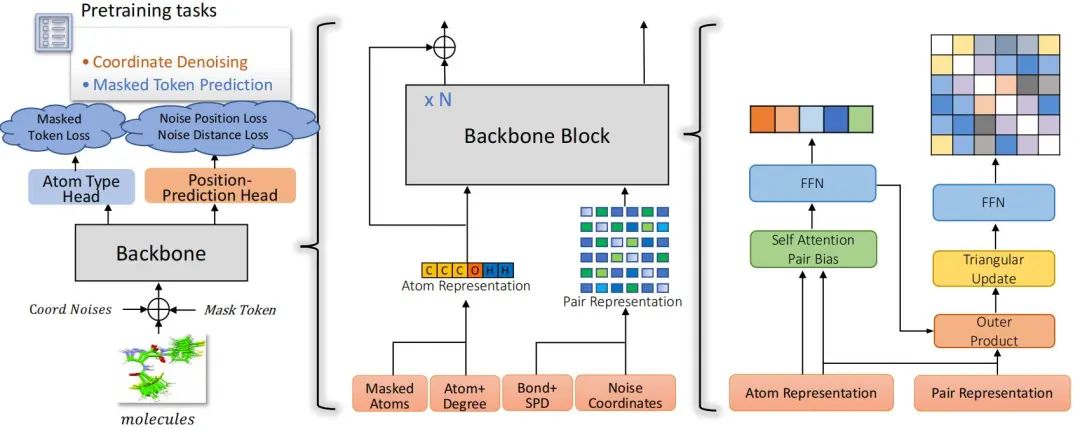
Figure 1: The overall pretraining architecture and details of backbone block
Uni-Mol2 uses a dataset containing 885 million compounds, 40 times the size of the dataset used in Uni-Mol training, with a 17-fold increase in the number of molecular skeletons, greatly expanding the data diversity.
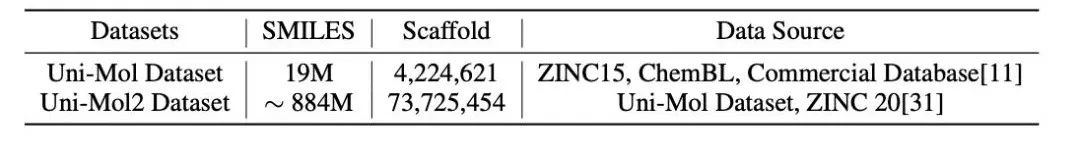
Table 1: Uni-Mol2 vs Uni-Mol Dataset Scale Comparison
Based on this, Uni-Mol2 shows better prediction performance. On the QM9 dataset for quantum mechanical prediction tasks, Uni-Mol2 achieves an average 27% performance improvement. When using the COMPAS-1D dataset, the 1.1-billion-parameter model of Uni-Mol2 achieves an average 4% performance improvement compared to Uni-Mol, and when the 1.1-billion-parameter model of Uni-Mol2 includes atomic and bond features, it achieves an average 14% performance improvement compared to Uni-Mol.
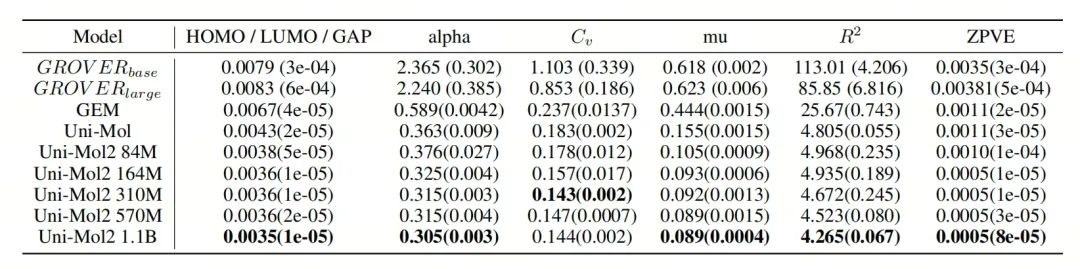
Table 2: The results of the mean absolute error [MAE, ↓] of the QM9 dataset
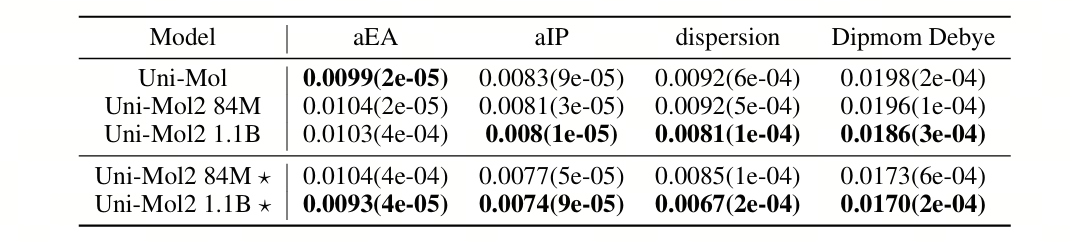
Table 3: The results of the mean absolute error [MAE, ↓] of the COMPAS - 1D dataset
Uni-Mol2 Scale Law Introduction
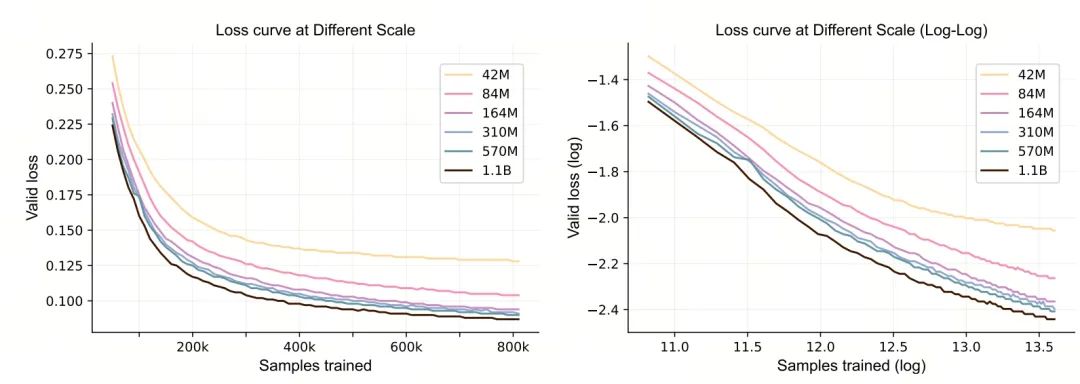
The research systematically explores the scale law in molecular pre-training. The experimental results show that during the training process of Uni-Mol2, as the parameter scale increases, the validation loss continuously decreases. Further analysis reveals that as the model scale expands, the relationship between the magnitude of the loss decrease and the molecular data scale, model parameter scale, and total computational scale can be characterized by a power law. This finding provides an important experimental basis for constructing larger-scale molecular pre-training models in the future.
Uni-Mol Tools Support Uni-Mol2
Compared with Uni-Mol, Uni-Mol2 has a good gain in tasks strongly related to the molecular ground state conformation, such as quantum chemical property prediction. For the convenience of users, currently, the code of Uni-Mol2 has been integrated into Uni-Mol Tools. Through the following few lines of code, multiple versions of Uni-Mol2 can be called through Uni-Mol Tools. Currently, it supports loading the weights of all release versions of the foundation model, and users can try different scale weights in multiple scenarios.
# Install unimol_tools
pip install unimol_tools --upgrade
# Uni-Mol2 is introduced in the version of unimol_tools==0.1.1, please note the installed version number
Examples of using Uni-Mol2 for training and prediction are as follows:
# In MolTrain and UniMolRepr, set model_name to unimolv2
# trainingfrom unimol_tools import MolTrain, MolPredict
clf = MolTrain(task='regression',
data_type='molecule',
epochs=100,
batch_size=4,
split='random',
save_path='./exp',
remove_hs=False,
early_stopping=5,
target_cols='TARGET',
model_name='unimolv2', # avaliable: unimolv1, unimolv2
model_size='84m', # work when model_name is unimolv2. avaliable: 84m, 164m, 310m, 570m, 1.1B.
)
clf.fit(data='path/to/train/file')
# inference
clf = MolPredict(load_model='./exp')
res = clf.predict(data='path/to/test/file')
The features of Uni-Mol2 can be obtained as follows:
# unimol2 representation
from unimol_tools import UniMolRepr
# single smiles unimol representation
clf = UniMolRepr(data_type='molecule',
remove_hs=False,
model_name='unimolv2', # avaliable: unimolv1, unimolv2
model_size='84m', # work when model_name is unimolv2. avaliable: 84m, 164m, 310m, 570m, 1.1B.
)
smiles_list = ['O=C1NCC1C1CC1']
unimol_repr = clf.get_repr(smiles_list, return_atomic_reprs=True)
Visit the tutorial for more information: https://unimol.readthedocs.io/en/latest/quickstart.html
Quick Access Address
Uni-Mol in the DeepModeling community repository: https://github.com/deepmodeling/Uni-Mol
Uni-Mol users and developers' questions: https://github.com/deepmodeling/Uni-Mol/issues
Uni-Mol Tools English documentation: https://unimol.readthedocs.io/en/latest/
Project Contribution
Xiaohong Ji, Zhen Wang, Zhifeng Gao, Hang Zheng, Yaning Cui, Letian Chen, Linfeng Zhang, Guolin Ke, Weinan E have contributed to the above work.