What Can Uni-Mol Do Too? | Implementing the Multimodal Alignment Encoder MolBind to Facilitate Drug Discovery
In current biological and chemical research, multimodal learning has become a crucial tool for advancing drug discovery and molecular understanding. However, most existing pre-training frameworks are limited to two modalities, and designing a unified network capable of handling multiple modalities such as natural language, 2D molecular graphs, 3D molecular conformations, and 3D proteins remains challenging. To address this issue, researchers from Pennsylvania State University and Tsinghua University proposed MolBind, a framework that trains multimodal encoders through contrastive learning and maps all modalities to a shared feature space. Uni-Mol plays a key role in this paper. As the core model for 3D conformation encoders and 3D protein encoders, it can effectively capture the 3D structural information of molecules and proteins, providing important support for MolBind to achieve accurate multimodal alignment. The preprint of the research "MOLBIND: Multimodal Alignment of Language, Molecules, and Proteins" was published on arXiv (https://arxiv.org/pdf/2403.08167).
Research Background
In recent years, significant progress has been made in multimodal pre-training technology in the fields of computer vision and natural language processing. The CLIP model, through the contrastive learning mechanism, aligns images and text and demonstrates powerful capabilities in cross-modal understanding and zero-shot tasks. This successful paradigm has inspired the exploration of more complex data modalities, especially in the biological and chemical fields. Scientific research data in these fields not only include text descriptions but also various modalities such as 2D molecular structures, 3D molecular conformations, and 3D protein structures.
However, existing methods mainly focus on single modality pairs (such as molecule-language or molecule-protein), making it difficult to expand to more complex multimodal tasks and limiting the generalization ability of models. In addition, the available molecule-language data in the biomedical field is much less than image-text data. Currently, there are only about 300,000 molecule-language pairs, which is several orders of magnitude smaller than the 400 million image-text pairs used for CLIP training. This data shortage restricts the application of multimodal pre-training in the biological and chemical fields, especially in tasks such as molecular generation, cross-modal retrieval, and protein-molecule interaction prediction.
To address these challenges, MolBind emerged. MolBind is a multimodal pre-training model that spans text, 2D molecules, 3D molecules, and protein structures. By aligning the data distributions of different modalities (Figure 1), MolBind constructs a unified cross-modal representation, which can support tasks such as language-guided molecular generation, cross-modal retrieval, and protein-molecule interaction modeling, making up for the limitations of existing methods and providing a more powerful AI-empowered solution for the biomedical field.
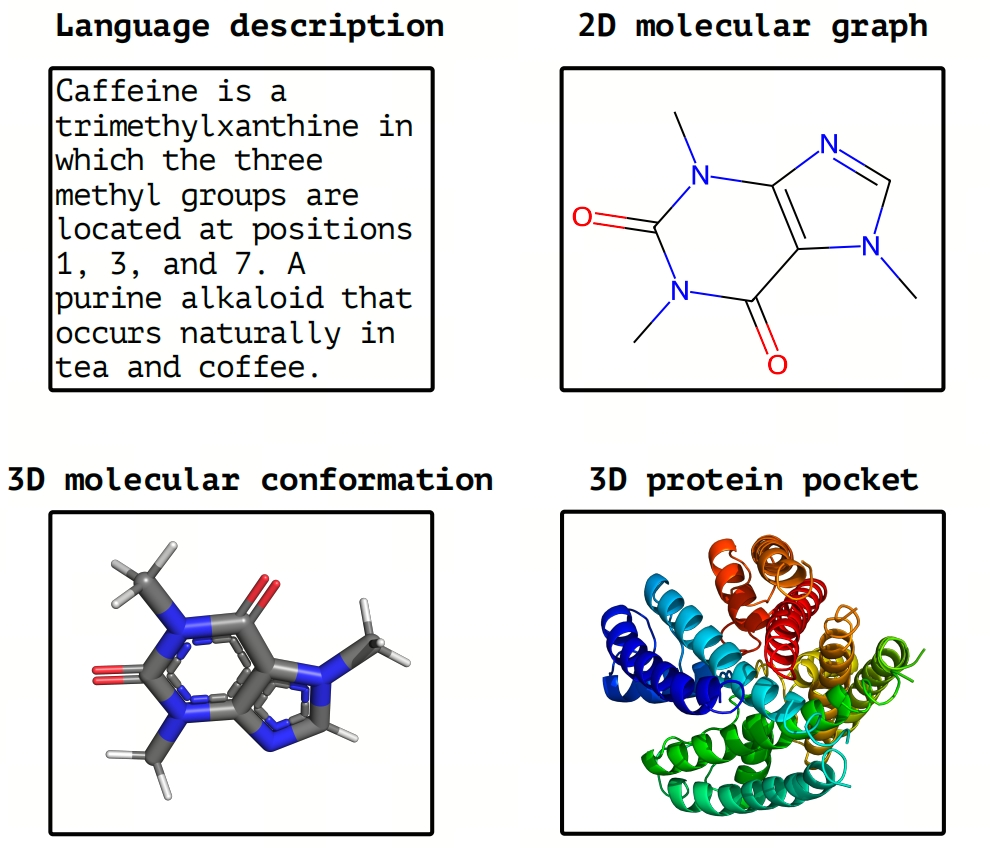
Figure 1: Illustration of modality data we try to align in this paper. The scientific language: text describes the name, property, and structure of molecules. 2D molecular graph: the atoms and bonds are nodes and edges in the graph. 3D molecular conformation: the atoms in the 3D Euclidean space. 3D protein pocket: a specific 3D region on the surface of a protein that can bind or interact with other molecules.
Method Introduction
The MolBind framework aligns multiple modalities (text, 2D molecules, 3D molecules, protein structures) into a unified embedding space through contrastive learning. Specifically, MolBind uses the following encoders:
Language Encoder: SciBERT is used to encode biomedical texts. SciBERT is a language model pre-trained on biological text data, which can effectively extract semantic information about molecular structures, properties, and functions from the text, providing accurate language feature representations for subsequent multimodal alignment.
2D Graph Encoder: A pre-trained Graph Isomorphism Network (GIN) is used to encode 2D molecular graphs. GIN is pre-trained through graph contrastive learning on 2 million molecules from ZINC15, which can accurately capture the structural information of atoms and bonds in 2D molecular graphs and convert it into feature vectors suitable for multimodal fusion.
3D Conformation Encoder: The Uni-Mol model is adopted to encode 3D molecular conformations. Uni-Mol is a Transformer-based model pre-trained on 209 million molecular conformations. It can effectively capture the 3D geometric information of molecules, precisely representing the shape of molecules in three-dimensional space and the relative positional relationships between atoms, providing key 3D structural features for multimodal fusion.
3D Protein Encoder: The Uni-Mol model is also used to encode 3D protein pockets. Initialized with the checkpoint pre-trained on a dataset of 3 million candidate protein pockets, the model can learn the structural features of protein pockets and provide key protein-related information for multimodal alignment.
similarity of paired data and minimize the similarity of unpaired data. Specifically, given a sample $x_{i}$ and its corresponding observation $y_{i}$ in another modality, MolBind encodes them into normalized embedding representations, denoted as $x_{i}=f_{\mathcal{X}}(x_{i})$ and $y_{i}=f_{\mathcal{Y}}(y_{i})$ respectively, where $f_{\mathcal{X}}$ and $f_{\mathcal{Y}}$ are the encoders corresponding to modalities X and X. Then, it is optimized through the following contrastive loss function:
$$
L_{\mathcal{X} 2 \mathcal{Y}} = -\log\frac{\exp\left(x_{i}^{\top} y_{i} / \tau\right)}{\exp\left(x_{i}^{\top} y_{i} / \tau\right)+\sum_{j \neq i} \exp\left(x_{i}^{\top} y_{j} / \tau\right)}
$$
where $\tau$ represents the temperature, and $y_{j}$ represents the negative samples within the batch. MolBind uses the symmetric loss $L_{\mathcal{X} 2 \mathcal{Y}}+L_{\mathcal{Y} 2 \mathcal{X}}$ to align X and Y, and this loss function is used to jointly bind the four modalities (language-graph, language-conformation, graph-conformation, and conformation-protein).
Dataset Construction
To effectively pre-train MolBind, we constructed a high-quality multimodal dataset MolBind-M4, including language-graph, language-conformation, graph-conformation, and conformation-protein paired data. The specific data sources are as follows:
Language-Graph Structure Pairs: Molecules and their text descriptions were obtained from PubChem, with a total of 322K pairs. Molecular annotations were downloaded using Pug View, and molecules were converted into graph structures and paired with text descriptions. To ensure the learning effect of the model, some descriptions were uniformly processed, and high-quality validation and test sets were also divided.
Language-Conformation Pairs: Molecular conformations were obtained from the Molecule3D and GEOM datasets and matched with text descriptions in PubChem, with a total of 161K pairs. Since traditional methods for generating molecular conformations have problems with accuracy and generalization, ground-state conformation data was selected, and conformations and text descriptions were matched through different identifiers.
Conformation-Protein Pairs: Conformation-protein pairs were extracted from the PDBBind and CrossDocked datasets, with a total of 72K pairs. After data screening, clustering, and other processing, data that did not meet the requirements was removed, and finally, a high-quality dataset was formed. The CASF-2016 dataset was used as the test set.
Graph-Conformation Pairs: The 3D molecular structures in the language-conformation dataset were directly converted into 2D graph structures, with a total of 161K pairs.
Research Results
MolBind performs well in multiple downstream tasks, including zero-shot molecule-language retrieval, zero-shot classification, and molecule-protein retrieval. The specific results are as follows:
- Zero-Shot Molecule-Language Retrieval: MolBind significantly outperforms existing methods in the "language-graph" and "language-conformation" retrieval tasks. See Table 1 below, which shows the performance comparison of different models in cross-modal retrieval tasks. MolBind performs outstandingly in various indicators. Using recall rate as the evaluation index, MolBind far exceeds the baseline models in both in-batch retrieval and full test set retrieval, demonstrating its effectiveness in aligning molecule and language modalities.
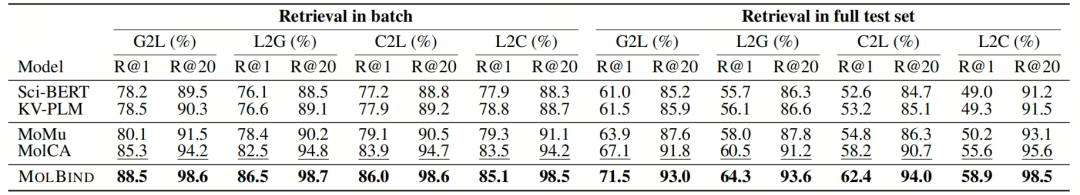
Table 1: Performance of cross-modal retrieval on test sets of MolBind-M4 includes the following scenarios: Graph to Language (G2L), Language to Graph (L2G), Language to Conformation (L2C), and Conformation to Language (C2L) retrieval.
- Zero-Shot Classification: MolBind performs remarkably in the IUPAC naming classification task. Figure 2 shows the results of the zero-shot molecular classification experiment. MolBind significantly outperforms the baseline models in this task. The IUPAC naming system relies on the accurate identification of molecular structures. MolBind can accurately predict IUPAC names by virtue of its aligned representation space of molecules and language descriptions, demonstrating its powerful ability to understand molecular structures.
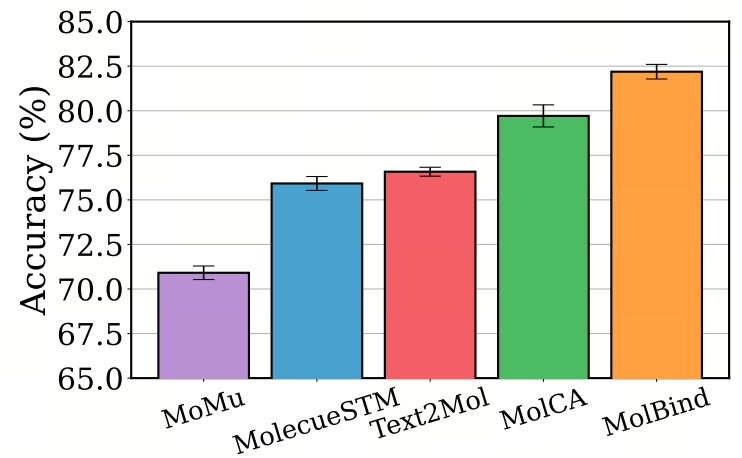
Figure 2: Zero-shot molecule classification with language.
- Molecule-Protein Retrieval: MolBind significantly outperforms Vina, Glide, and DrugCLIP in the conformation-protein retrieval task. Table 2 presents the recall rate comparison of different models in the conformation-protein retrieval task. MolBind has a significantly higher recall rate. This task is of great significance in drug discovery, and the excellent performance of MolBind verifies its ability to align molecular conformations and protein modalities using multiple modalities.
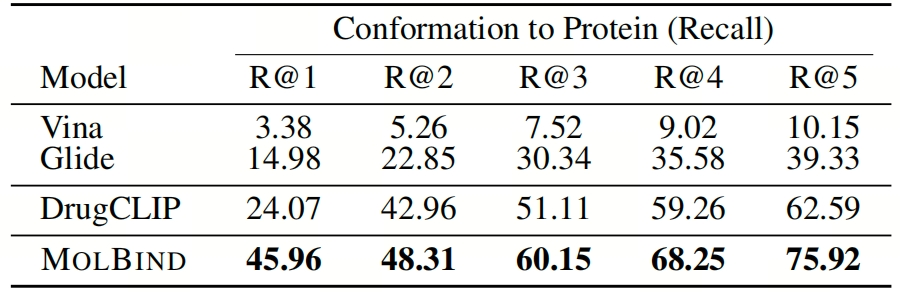
Table 2: Conformation-to-protein retrieval on CASF-2016.
Summary
MolBind is a simple and effective framework that can achieve multimodal alignment in the context of insufficient biochemical data and promote knowledge transfer between modalities. The article also constructed the multimodal dataset MolBind-M4, which is the first multimodal dataset completely sourced from public data. The outstanding performance of MolBind in multiple downstream tasks verifies its effectiveness in multimodal representation alignment, providing a new and powerful tool for biological and chemical research and promising to further advance drug discovery, molecular understanding, and other aspects.