What Can Uni-Mol Do Too? | Facilitating AI-Powered Design of Lipid Nanoparticles
In recent years, with the rapid advancement of mRNA vaccines and nucleic acid drugs, lipid nanoparticles (LNPs) have emerged as one of the most crucial drug delivery tools. However, the performance of LNPs depends on various lipid components and their proportions. Experimental optimization is not only time-consuming and labor-intensive but also struggles to cover the vast design space. Recently, the work led by Alvin Chan and his team was published in Nature Nanotechnology under the title "Designing lipid nanoparticles using a transformer-based neural network". The study proposes a transformer-based neural network model called COMET, which integrates molecular structures and formulation parameters to predict LNP performance. A key component of this process is the use of Uni-Mol as the core tool for molecular representation learning.
Challenges in LNP Design
The core of an LNP consists of four types of lipids: ionizable lipids, cholesterol, helper lipids, and PEG lipids. These molecules not only have complex structures themselves but also exhibit vastly different performances under varying proportions and mixing conditions. For instance, altering the N/P ratio (nitrogen-to-phosphorus ratio) or adjusting the mixing ratio of organic and aqueous phases can significantly impact transfection efficiency (see Figure 1). In such a highly multi-factor coupled system, it is nearly impossible to exhaust all possible combinations solely through experiments.
To address this, the research team constructed an unprecedentedly large experimental dataset named LANCE. This dataset systematically collects data on over 3,000 LNP formulations and their transfection efficiencies in mouse cells. In addition to conventional single-ionizable lipid combinations, the dataset also includes dual-ionizable lipids, different cholesterol derivatives, and polymeric materials, providing abundant training materials for the AI model.
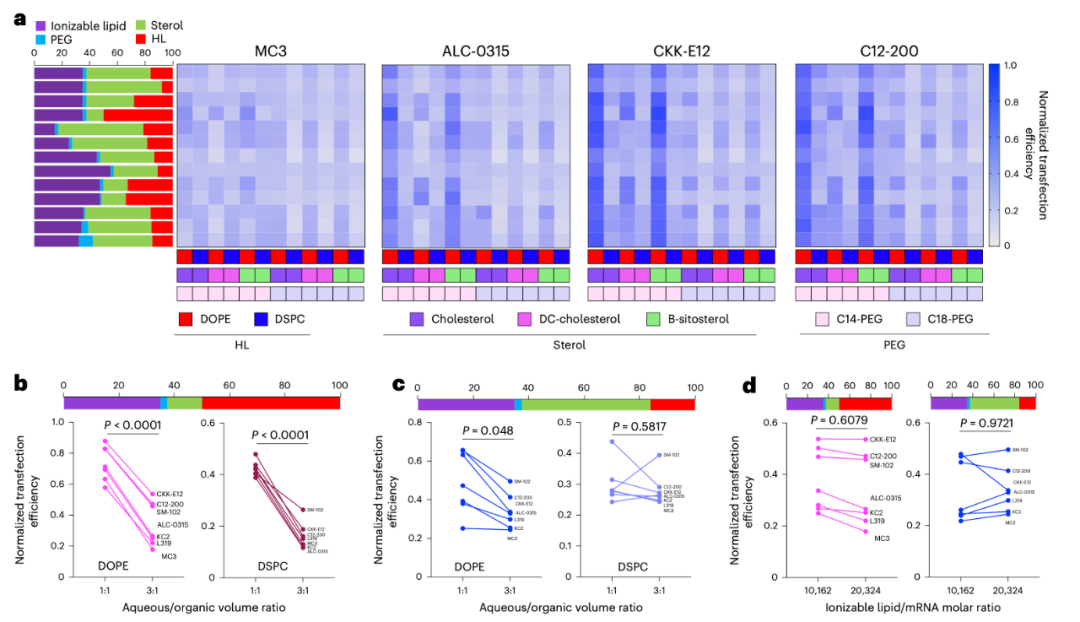
Figure 1: a. Effect of lipid selection and proportion on the transfection efficiency of LNPs in DC2.4 cells (HL stands for helper lipid).b-c. Effect of aqueous/organic phase volume ratio on the transfection efficiency of LNPs with a high proportion of helper lipids (b) and a low proportion of helper lipids (c) in DC2.4 cells.d. Effect of ionizable lipid/mRNA weight ratio on the transfection efficiency in DC2.4 cells.
The COMET Model: Understanding Formulations Like a "Language Model"
Building on this foundation, the research team designed the COMET (Composite Material Transformer) model. The unique feature of this model lies in treating each lipid molecule as a "token" and encoding different proportions and experimental parameters (such as N/P ratio and mixing ratio) into "tokens" as well, which are ultimately fed into the transformer model for modeling (see Figure 2).
The Uni-Mol model (a general 3D molecular representation learning framework) is employed in this process, which can convert the 3D structure of each lipid molecule into a vector representation. Unlike traditional models that rely on handcrafted features, Uni-Mol can directly extract information from atomic coordinates, enabling the COMET model to "interpret" complex molecular structures. It can be said that without Uni-Mol, seamlessly integrating information at the molecular structure level into the formulation prediction process would be extremely challenging.
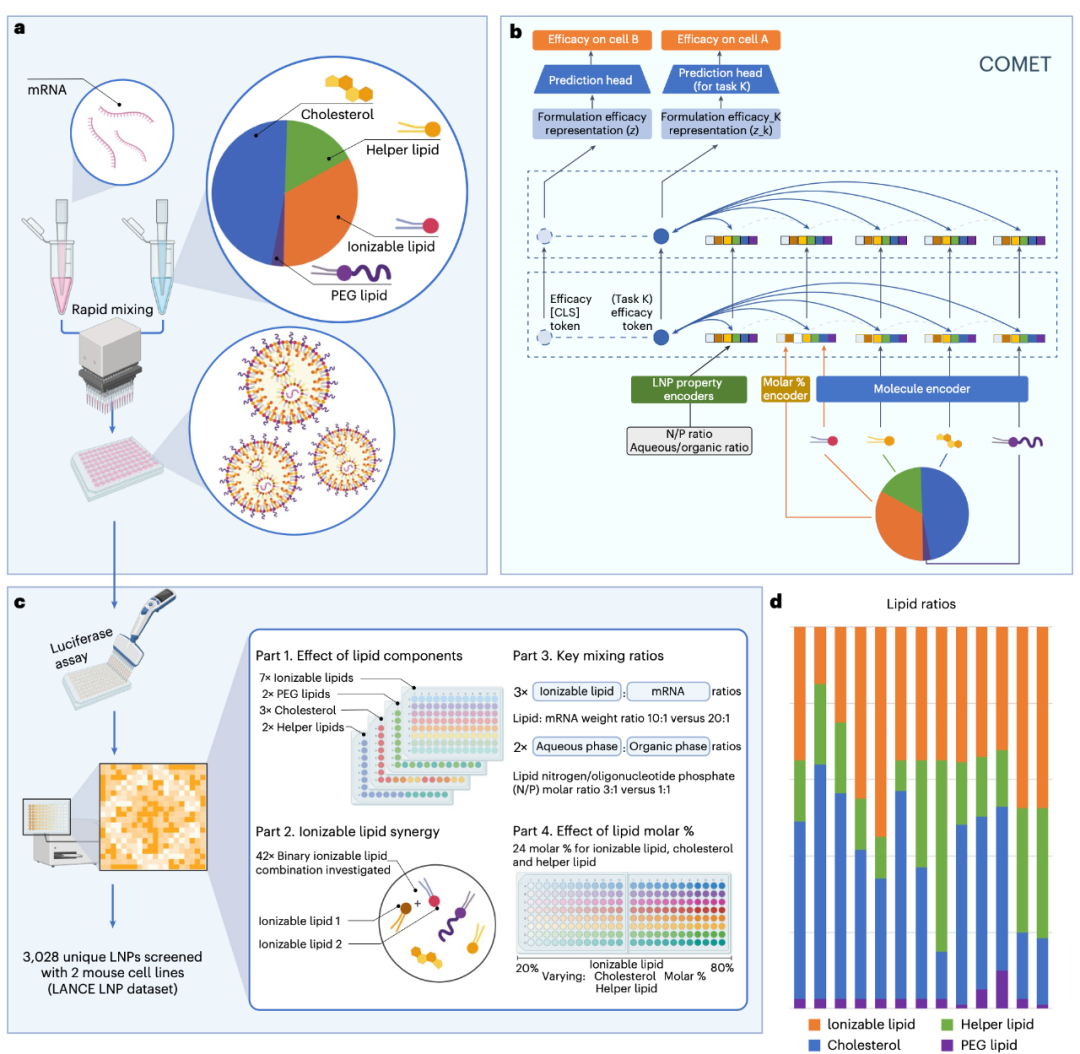
Figure 2:a. The synthesis of LNPs is achieved by mixing nucleic acids (e.g., mRNA) with a lipid solution that typically contains four types of lipids. Their key properties (such as transfection efficiency) depend not only on the lipid structure but also are closely related to the relative proportions of each component and mixing parameters (e.g., N/P ratio, aqueous/organic phase volume ratio).b. The COMET platform can predict the performance of composite materials based on component materials (e.g., lipids composing LNPs), proportion parameters, and other conditions.c. Through high-throughput screening technology, the training data of COMET covers four complementary LNP formulation space modules.d. Thirteen main lipid molar proportion schemes used in the training dataset.
From Prediction to Discovery: AI Screening for Novel LNPs
The COMET model performed exceptionally well on the LANCE dataset, accurately predicting LNP efficacy with a Spearman correlation coefficient close to 0.9. More excitingly, the model can conduct exploration in a "virtual space" — the research team used it to screen nearly 50 million virtual LNP combinations, ultimately selecting dozens of high-scoring candidate formulations, which were then verified through experiments (see Figure 3). The results showed that these "AI-discovered" formulations not only outperformed clinically approved LNPs (such as SM-102 and MC3) in in vitro experiments but also demonstrated stronger mRNA delivery capabilities in in vivo mouse experiments.
In this process, Uni-Mol played a crucial role: it provided reliable molecular structure representations for the COMET model, enabling the model to "distinguish" subtle differences between different lipids and thereby discover novel formulations that are difficult to predict using traditional methods.
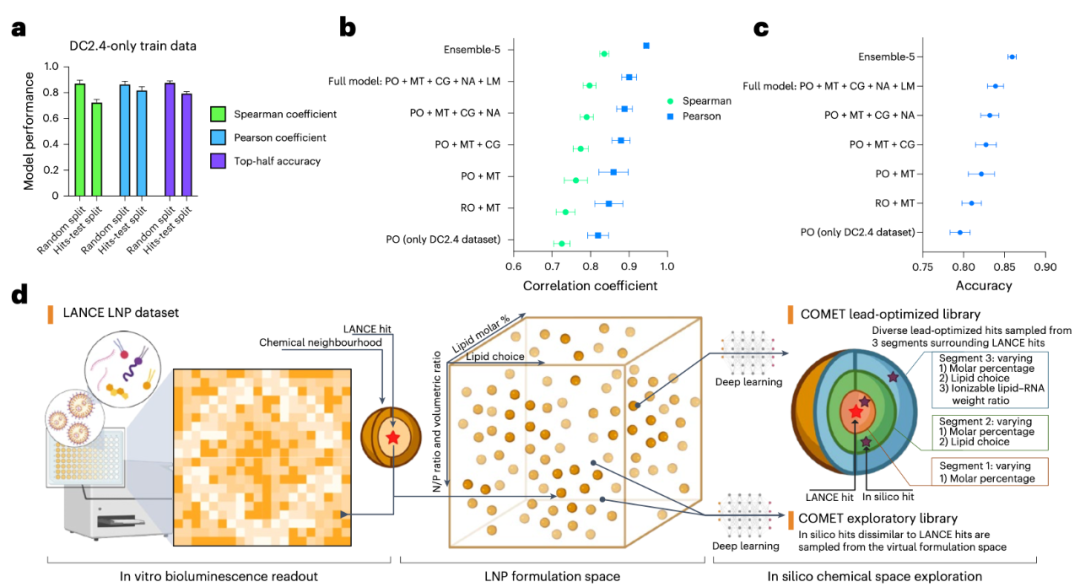
Figure 3: a. Performance of COMET under different test dataset partitions after training on LNP efficacy data from DC2.4 cells.b-c. Results of ablation experiments showing the contribution of each module to the ranking performance (b) and prediction accuracy (c) of COMET in the 'hits-test' dataset of DC2.4 cells.d. Schematic diagram of the computer-aided screening process: starting from a large virtual LNP library, virtual screening is conducted via COMET, followed by filtering based on properties such as efficacy and diversity.Abbreviation notes: MT = Multi-task Learning, RO = Regression Objective, PO = Pairwise Ordering Objective, CG = CAGrad Algorithm, NA = Noise Augmentation, LM = Label Margin.
Beyond LNPs: The Generalization Ability of the Model
Furthermore, the researchers tested the scalability of the COMET model. The results showed that the model can not only handle traditional LNPs but also be extended to the following scenarios:
- Dual-ionizable lipid formulations: It can capture the synergistic effects between lipids, significantly improving delivery efficiency;
- Novel polymeric materials (e.g., PBAEs, poly(β-amino esters)): It can incorporate entirely new structures such as poly(β-amino esters) into modeling and successfully optimize formulations with higher efficacy (see Figures 4a and 4b);
- New cell types and new RNA cargos: For example, predicting the mRNA delivery efficiency in human-derived Caco-2 cells and HepG2 cells (see Figures 4j-m);
- Lyophilization stability: It can predict the efficacy decay of LNPs after lyophilized storage, providing references for the practical application of drugs (see Figures 4n-o).
These results fully demonstrate that relying on the molecular representations provided by Uni-Mol, the COMET model possesses strong adaptability and scalability, truly enabling AI to participate in the "full-process" design of drug delivery systems.
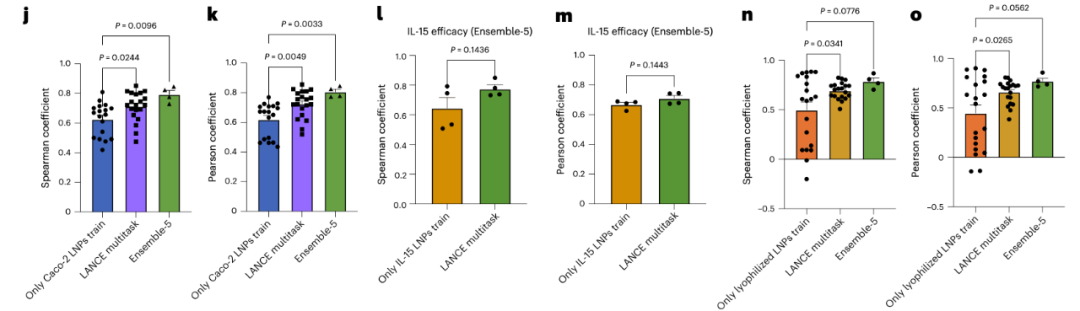
Figure 4: a. Structural characteristics of branched PBAEs (poly(β-amino esters)).b. Strategy for integrating PBAE and LNP properties in the inference process of COMET.j-k. Performance evaluation of COMET in predicting the efficacy of LNPs in Caco-2 cells.l-m. Performance evaluation of COMET in predicting the IL-15 mRNA delivery efficacy in HepG2 cells.n-o. Performance evaluation of COMET in predicting the efficacy decay of LNPs after lyophilization.Except for Ensemble-5, which used 4 replicates, the evaluations in j-o all adopted 20 replicate experiments. Error bars represent standard errors. For j, k, n, and o, one-way ANOVA with post-hoc Dunnett’s test was used; for l-m, unpaired two-tailed t-tests were used.
Conclusion
This study demonstrates the role of deep learning in addressing the complex formulation design challenges in the field of drug delivery. With the 3D molecular representations provided by Uni-Mol, the COMET model can not only comprehensively integrate molecular structures and formulation parameters but also quickly screen out truly efficient and scalable candidate systems from the vast combinatorial space. This approach provides a new paradigm for the development of nucleic acid drugs and vaccines, and also highlights the core value of AI in future drug design and material innovation.