UniMol_Tools v0.15: Open-Source Lightweight Pre-Training Framework for One-Click Reproduction of Original Uni-Mol Accuracy!
The official release of UniMol_Tools v0.15 introduces lightweight pre-training and a synchronized full-process command-line tool based on Hydra. Developers can complete the entire workflow from preprocessing → pre-training → fine-tuning → property prediction with just a few lines of code, and the reproduced results are nearly identical to those of the original Uni-Mol. This new version aims to provide an efficient and reproducible computing platform for research in materials science, medicinal chemistry, and molecular design.
Core Highlights
This release marks the first research tool on the market that simultaneously covers molecular representation, property prediction, and custom pre-training, offering an efficient and reproducible computing platform for studies in materials science, medicinal chemistry, and molecular design.
- Lightweight Pre-Training
The complete pipeline supports masking strategies, multi-task loss functions, metric aggregation, and distributed training, while being compatible with custom pre-trained models and dictionary paths. - One-Command Execution
Hydra configuration management enables one-click execution of training, representation, and prediction workflows, making experimental reproduction more efficient. - Research-Friendly Optimizations
Features dynamic loss scaling, mixed-precision training, distributed support, and checkpoint resumption, adapting to large-scale molecular data. - End-to-End Modeling
Provides a one-stop solution for data preprocessing, model training, molecular representation generation, and property prediction. - Extensibility & Configurability
Offers abundant configuration files and examples for quick onboarding and customization of personalized tasks.
Comparison Between UniMol_Tools v0.15 and the Original Uni-Mol
| Capability | This Release | Original Uni-Mol |
|---|---|---|
| Pre-training Code Lines | Newly written, over 2,000 lines | Over 6,000 lines |
| Distributed Training | Natively supports DDP & mixed precision | Requires manual configuration |
| Data Formats | csv / sdf / smi / txt / lmdb | Only lmdb |
| Downstream Fine-Tuning | Weight zero conversion; direct use of unimol_tools.train/predict | Requires manual format modification |
One-Command Pre-Training
The new version delivers an "out-of-the-box" training experience. Research users can complete the entire pre-training workflow from data preprocessing to model training with a single command, significantly lowering the barrier to experimentation.
1 | torchrun \ # DDP |
Technical Details
- Multi-Target Masking Loss (Masked Token + 3D Coord + Dist Map)
The pre-training curve overlaps with the original Uni-Mol by over 99%, ensuring stable performance.
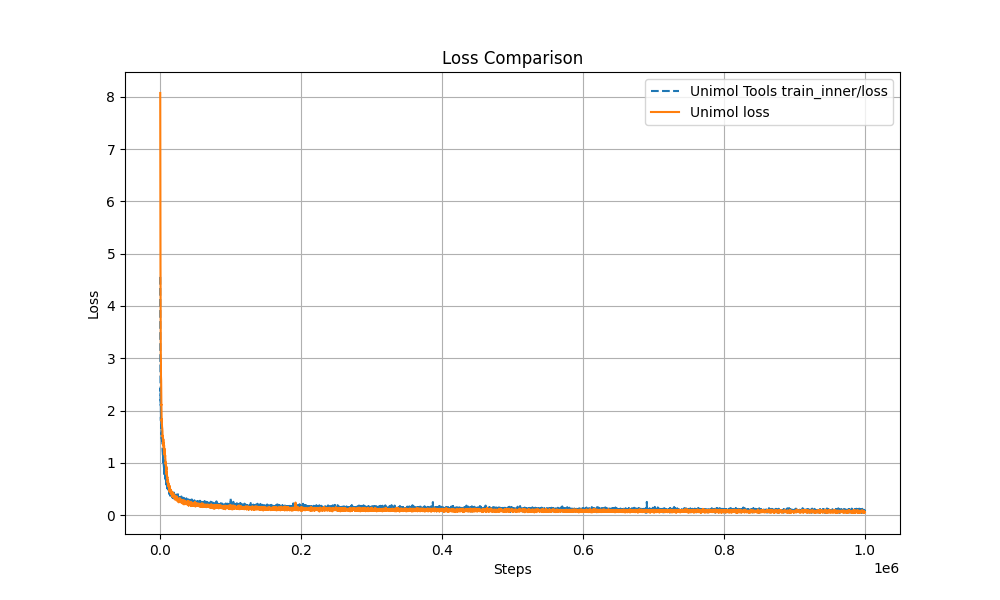
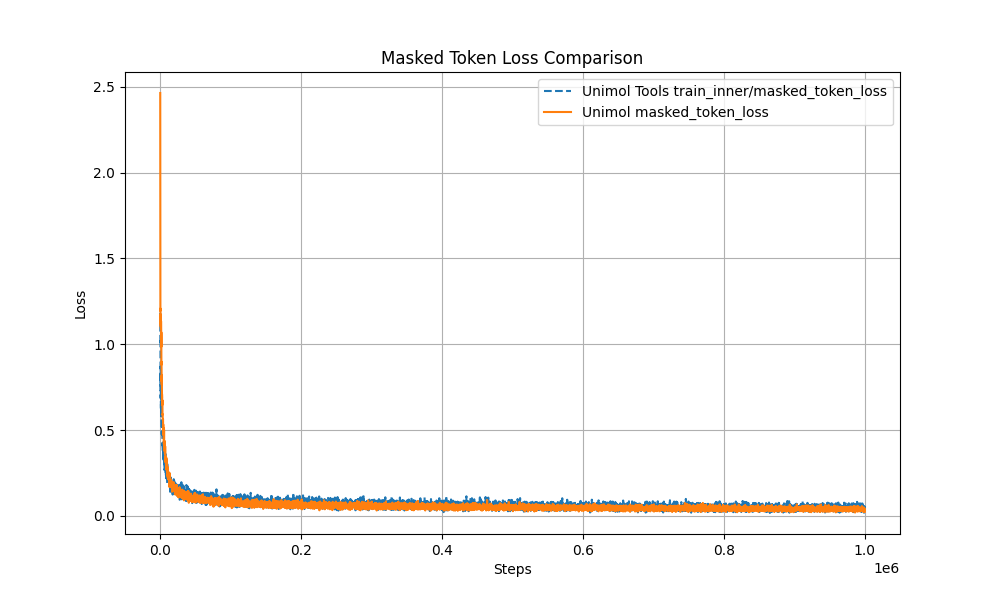
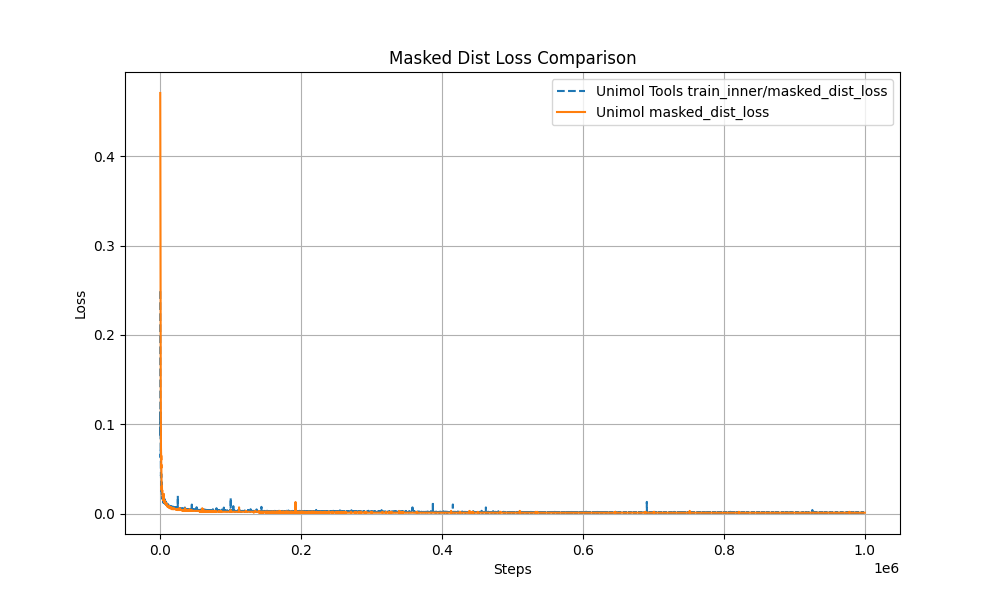
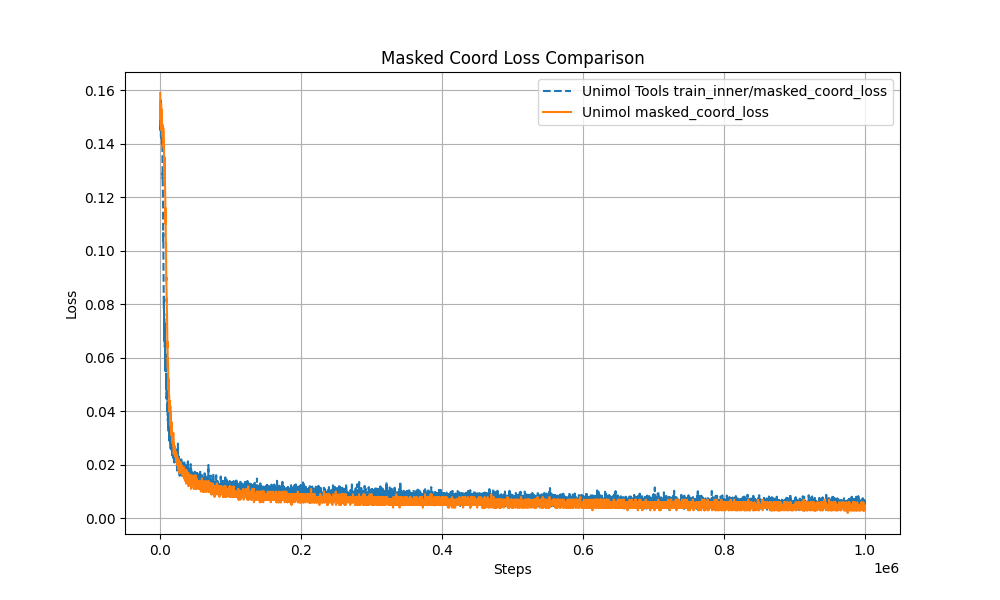
- Modular Design
The complete workflow can be reproduced with just four files:
1 | unimol_tools/pretrain/ |
This minimizes the threshold for secondary development—modify just one line of configuration to run custom tasks.
- Backward Compatibility
- Existing APIs such as unimol_tools.train / predict / repr remain unchanged;
- Supports passing custom pretrained_model_path and dict_path—old scripts only need two additional parameters to load new weights;
Overvoew of Updates
- Lightweight pre-training module: The complete pipeline supports masking strategies, multi-target loss for 3D coordinates and distance matrices, metric aggregation, and distributed training;
- Hydra full-process CLI: One command to run training, representation, and prediction; parameters can be quickly adjusted;
- Enhanced data processing: Supports csv / sdf / smi / txt / lmdb, flexibly adapting to formats commonly used by research users;
- Optimized distributed training: Native DDP + mixed precision, supporting checkpoint resumption;
- Modular design: The complete workflow can be reproduced with only four core files, facilitating secondary development;
- Compatibility with old-version APIs: Load new pre-trained weights without modifications, supporting custom models and dictionary paths;
- Performance and reproducibility guarantee: Pre-training curve is highly consistent with the original Uni-Mol;
Open-Source Community
UniMol_Tools is one of the open-source projects under the DeepModeling community. Developers interested in the project are welcome to participate long-term:
- GitHub Repo: https://github.com/deepmodeling/unimol_tools
- Documentation: https://unimol-tools.readthedocs.io/
- The Issue section welcomes feedback on problems, suggestions, and feature requests;
- New users can refer to the Readme and documentation for quick onboarding.
If you encounter any issues during use, please submit an Issue on GitHub or contact us via email.
About Uni-Mol
Uni-Mol is a widely acclaimed molecular pre-training model in recent years, dedicated to building a universal 3D molecular modeling framework. As its derivative toolkit, UniMol_Tools aims to lower the application threshold of the model and improve development efficiency.