DeePMD-kit v3 Tutorial 3 | Model Plugin Mechanism and Training MACE Models
The third highlight of DeePMD-kit v3 is its model plugin mechanism. This article introduces the feature from three perspectives: background and principles, development tutorial, and usage tutorial.
Background and Basics
Currently, most deep learning potential model software only supports the potential models developed by their authors. In other words, when developing potential models, the authors “incidentally” develop a set of software. DeePMD-kit also evolved from the Deep Potential series of models. This brings about several issues:
High learning costs: Users need to learn a new set of software for each new model, whereas different DFT functionals can generally be used within the same software.
High development costs: Model developers not only need to write the model but also develop and maintain interfaces for training models, connecting with MD software, etc.
Difficulty in fair comparisons: It is challenging to compare different model structures fairly because the loss functions, learning rates, and data processing methods of different software may vary greatly.
Therefore, DeePMD-kit introduces a model plugin mechanism, allowing new or existing models to be integrated into DeePMD-kit as plugins. These models can then be connected to various MD software and invoked by workflow tools such as DP-GEN.
Development Tutorial
The repository https://github.com/njzjz/deepmd-gnn provides an example of a model plugin. It integrates MACE and NequIP models into DeePMD-kit, as illustrated in the figure below.
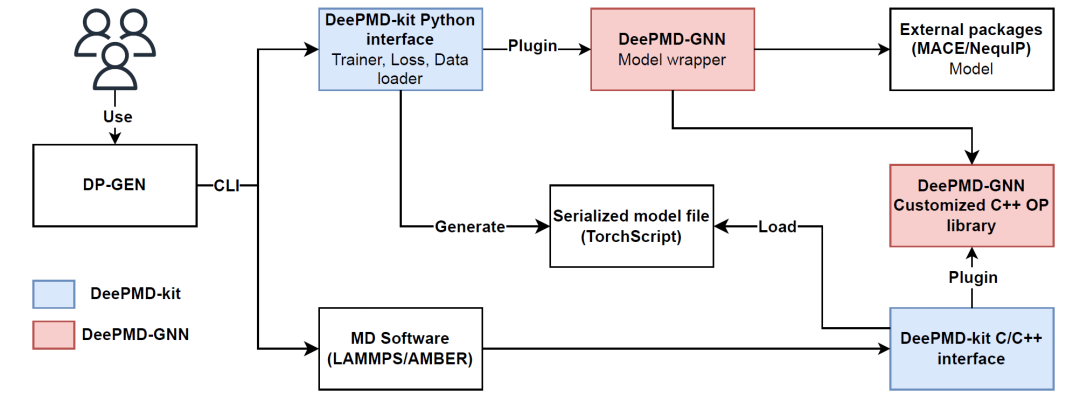
The most critical model wrapper code is shown as follows:
1 | from deepmd.pt.model.model.model import ( |
At the same time, it is necessary to register the parameters corresponding to the model:
1 | from dargs import Argument |
In the pyproject.toml file, register the model in the entry points specified by DeePMD-kit so that it can be automatically discovered:
1 | [project.entry-points."deepmd.pt"] |
Tutorial: Training a MACE Model
This tutorial can be run directly in a Notebook on Bohrium:
https://bohrium.dp.tech/notebooks/75755718691/
Training, Freezing, and Testing
1. Install the DeePMD-GNN Plugin
Follow the documentation to install the DeePMD-GNN plugin. Then, navigate to the example directory and modify the training steps to 2000:
1 | git clone https://github.com/njzjz/deepmd-gnn |
2. Example Model Configuration
The model section of the training script is defined by the plugin, while other parts (e.g., loss, learning rate, training) remain unchanged from previous tutorials. Here’s an example:
1 | "model": { |
3. Train, Freeze, and Test
Run the following commands to train, freeze, and test the model:
1 | dp --pt train input.json |
DP_GNN_USE_MAPPINGis a behavior defined by the plugin itself.
Running LAMMPS Simulations
1. Set Up the LAMMPS Environment
Switch to DeePMD-kit's LAMMPS example directory and copy the frozen model file:
1 | cd ../../../ |
2. Create a LAMMPS Input File
Create a file (e.g., mace.in) with the following content:
1 | units metal |
3. Set the Plugin Path and Run LAMMPS
Set the environment variable DP_PLUGIN_PATH to the custom operator library provided by the plugin, then run LAMMPS:
1 | export DP_PLUGIN_PATH=$CONDA_PREFIX/lib/python3.12/site-packages/deepmd_gnn/lib/libdeepmd_gnn.so |
Conclusion
By integrating the model into DeePMD-kit via the plugin mechanism, its usage becomes almost identical to native models, with only minor differences.
If you’ve developed your own model and wish to integrate it with DeePMD-kit, feel free to contact the DeePMD-kit development team on GitHub for guidance.