DeePMD-GNN: Integrating External Graph Neural Network Potentials into DeePMD-kit
Recently, the Journal of Chemical Information and Modeling published a research work titled "DeePMD-GNN: A DeePMD-kit Plugin for External Graph Neural Network Potentials" [1]. DeePMD-GNN is a plugin for DeePMD-kit [2], which for the first time achieves seamless integration of external Graph Neural Network (GNN) potential energy models within the DeePMD-kit framework, including mainstream models such as NequIP and MACE.
Previously, machine learning potential (MLP) models and corresponding software developed by different teams often suffered from poor interoperability: users had to continuously learn new software tools, while developers needed to individually create molecular dynamics (MD) interfaces for each model. DeePMD-GNN effectively addresses these issues. By leveraging DeePMD-kit’s flexible plugin mechanism, this plugin unifies the training and application interfaces for different GNN potential models. Furthermore, the plugin mechanism ensures fair performance comparisons between models. The paper conducts rigorous benchmark tests on mainstream potential models such as NequIP, MACE, and DPA-2 using the QDπ dataset, providing comprehensive and reliable comparative results.

DeePMD-GNN represents the first demonstration of DeePMD-kit’s plugin mechanism. In the future, this mechanism is expected to enable efficient integration of more diverse machine learning potential models into DeePMD-kit.
Paper URL: https://doi.org/10.1021/acs.jcim.4c02441
Performance Testing
After dividing the QDπ dataset into training and test sets, the table below shows the errors of different models on the test set after training with the same training set and number of steps. A comparison between pure MLP and SQM+ΔMLP (SQM=GFN2-xTB) is also included.
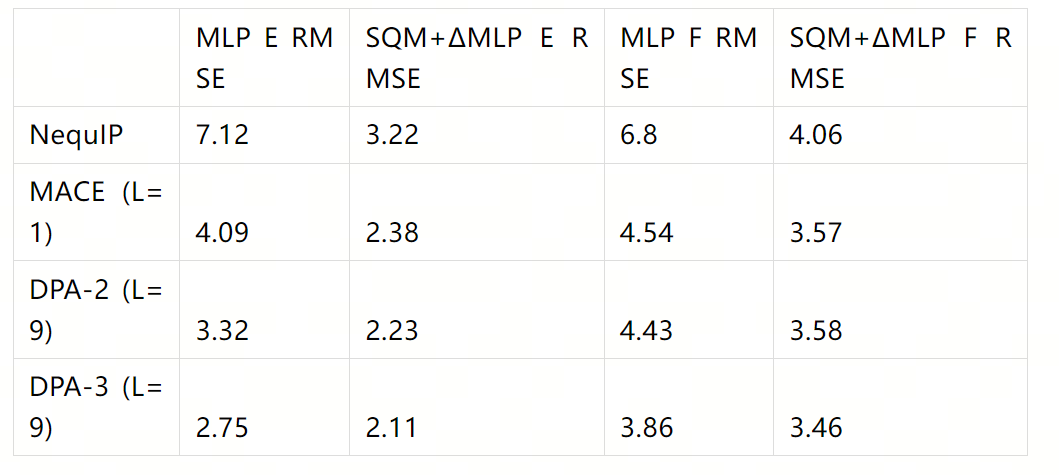
After dividing the QDπ dataset into training and test sets, the table below shows the errors of different models on the test set after training with the same training set and number of steps. A comparison between pure MLP and SQM+ΔMLP (SQM=GFN2-xTB) is also included.
Usage Instructions
The DeePMD-GNN software has been integrated into the DeepModeling open-source community:
https://github.com/deepmodeling/deepmd-gnn
One-click installation of DeePMD-GNN:
1 | conda install deepmd-gnn -c conda-forge |
Usage instructions for DeePMD-GNN were demonstrated in last year’s tutorial. Click https://mp.weixin.qq.com/s/FbS4uNPDWsZepQMQtOLj2Q to access.
Talent Recruitment
The first author of this paper, Jinzhe Zeng, is currently a tenure-track associate professor and doctoral supervisor at the School of Artificial Intelligence and Data Science, University of Science and Technology of China (major: 140500 Intelligent Science and Technology). He is recruiting outstanding undergraduates for doctoral studies.
Contact: jinzhe.zeng@ustc.edu.cn
References
[1] Jinzhe Zeng, Timothy J. Giese, Duo Zhang, Han Wang, Darrin M. York, "DeePMD-GNN: A DeePMD-kit Plugin for External Graph Neural Network Potentials," Journal of Chemical Information and Modeling, 2025, 65, 7, 3154-3160.
[2] Jinzhe Zeng, Duo Zhang, et al., "DeePMD-kit v3: A Multiple-Backend Framework for Machine Learning Potentials," Journal of Chemical Theory and Computation, 2025.