ReacNetGenerator | Partnering with the DeepModeling Community to Advance Research on Complex Reaction Systems
The ReacNetGenerator project has officially joined the DeepModeling open-source community, aiming to further advance its development.
Project Homepage
https://github.com/deepmodeling/reacnetgenerator
Background
In recent years, reactive molecular dynamics (Reactive MD) simulation methods have been widely used to study the reaction mechanisms of various complex molecular systems, such as:
- Combustion
- Explosions
- Heterogeneous catalysis
However, long-term MD simulations of large systems generate highly complex trajectory files that include numerous reactions and molecular species. Manually analyzing such trajectories is impractical.
About ReacNetGenerator
To address these challenges, the ReacNetGenerator software was introduced in the spring of 2018.
Key Features:
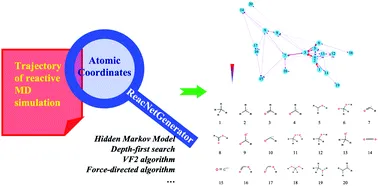
- Input Requirements: Uses atomic coordinates from the trajectory as its sole input.
- Automatic Analysis: Determines species and reactions based on atomic connectivity.
- Interactive Visualization: Presents reaction types, quantities, and networks on an interactive webpage.
- Users can click to analyze localized reaction networks involving specific species.
- Noise Filtering: Employs a Hidden Markov Model (HMM) to filter noise, highlighting key reactions.
- High Efficiency: Supports parallel computing and optimized memory usage, making it capable of analyzing large trajectory files.
Original Paper:
Jinzhe Zeng, Liqun Cao, Chih-Hao Chin, Haisheng Ren, John Z. H. Zhang, Tong Zhu,
ReacNetGenerator: an automatic reaction network generator for reactive molecular dynamics simulations,
Phys. Chem. Chem. Phys., 2020, 22 (2), 683–691.
https://doi.org/10.1039/C9CP05091D
Check out following publications employing ReacNetGenerator in their research: https://blogs.deepmodeling.com/papers/ReacNetGenerator/
Impact
By joining the DeepModeling community, ReacNetGenerator aims to further facilitate research on complex reaction systems, advancing our understanding of intricate molecular dynamics.
Installation and Usage of ReacNetGenerator
You can install ReacNetGenerator using either pip or conda:
Using pip:
1 | pip install reacnetgenerator |
Using conda:
1 | conda install reacnetgenerator -c conda-forge |
Verify Installation
After installation, you can check if ReacNetGenerator was installed successfully by running:
1 | reacnetgenerator -h |
If installed correctly, this will display the help information and available commands for the software.