What Can Uni-Mol Do Too? | Empowering ReactZyme to Reconstruct Biocatalytic Design and Overcome the Bottleneck of Enzyme Reaction Prediction
Enzymes are the core catalysts of life activities, and the prediction of their functions has important applications in various fields. However, traditional methods of enzyme classification, such as EC numbers or protein family classification, rely on manual classification, which can lead to inaccurate classification granularity. Moreover, these methods lack the ability to characterize the dynamic structural transformation of substrates and products during the reaction process, making it difficult to accurately understand the actual functions and catalytic mechanisms of enzymes. Against this backdrop, ReactZyme, developed by Hua et al., brings new hope to the study of enzyme functions. Uni - Mol, as the core engine for molecular modeling, provides powerful support for capturing the complex three - dimensional interactions between enzymes and substrates. This achievement was presented at the Datasets and Benchmarks track of the 38th Conference on Neural Information Processing Systems (NeurIPS 2024), and its code and data have been open - sourced(https://github.com/WillHua127/ReactZyme).
Research Background
The existing enzyme function annotation systems, EC (Enzyme Commission) and GO (Gene Ontology), are widely used. However, the manual - classification - based approach has two major drawbacks.
Firstly, inaccurate classification granularity affects the accurate determination of enzyme functions. EC mainly classifies enzymes according to the types of chemical reactions they catalyze and substrates. This may group enzymes with significantly different functions into the same category or over - segment enzymes with similar functions. For example, some enzymes with marked differences in catalytic mechanisms and reaction conditions are grouped together just because their substrates are similar. Although GO annotation is more comprehensive, it often lacks precise descriptions in specific function definitions, failing to meet the requirements of in-depth research.
Secondly, traditional methods have limitations in representing dynamic reactions. Enzyme catalysis is a dynamic process involving the binding of substrates to enzymes, the transformation of substrates, and the generation of products. Traditional enzyme function annotation methods struggle to capture the details of substrate - product structure transformation and cannot accurately reflect the actual catalytic mechanisms of enzymes in reactions. This restricts the in - depth understanding and accurate prediction of enzyme functions.
ReactZyme takes an alternative approach by directly establishing the mapping relationship between enzymes and catalytic reactions. Unlike traditional methods such as sequence alignment (e.g., BLAST) or EC - based prediction (e.g., CLEAN), ReactZyme uses machine learning techniques to deeply analyze reaction mechanisms and realizes the reverse prediction of "inferring enzyme functions from reactions". This innovative prediction method provides a new paradigm for the discovery of new enzymes and the annotation of enzyme functions, breaking through the limitations of traditional methods and opening up a new path for enzyme function research.

Figure 1: Overview of the enzyme - reaction prediction task. (a) Schematic diagram of the enzymatic reaction process: The substrate binds to the enzyme; an enzyme - substrate complex is formed; the product is released, enabling the enzyme to enter the next catalytic cycle. (b) Current methods for enzyme - reaction prediction: Search for annotated enzymes (e.g., sequence - based BLAST, structure - based FoldSeek); predict EC/GO annotations (e.g., CLEAN); enzyme - reaction prediction (ReactZyme).
Research Methods
Data Construction and Challenges
In data construction, the researchers carried out a large amount of meticulous and innovative work. They integrated the SwissProt and Rhea databases (data as of January 2024) to construct an ultra - large - scale dataset containing 178,327 enzymes and 7,726 reactions (Table 1). This dataset is much larger than traditional enzyme - reaction datasets such as ESP and EnzymeMap, providing a rich data foundation for subsequent research.
Table 1: Comparison of ESP, EnzymeMap, and ReactZyme
.png)
To address the problems of missing atom mapping and insufficient reaction coverage in traditional datasets, ReactZyme adopted multiple innovative strategies. The triple - split strategy of time, enzyme similarity, and reaction similarity is particularly crucial, as it can effectively simulate the prediction tasks of new enzymes and new reactions in real - world scenarios. In time - based splitting, the data is divided into training and test sets according to time nodes, enabling the model to learn the enzyme - reaction characteristics of different periods and enhancing its adaptability to newly emerging enzymes and reactions. Enzyme - similarity - based splitting calculates the Levenshtein distance of protein sequences to ensure that there are differences between the enzymes in the training and test sets, reducing the risk of overfitting and improving the model's generalization ability for new enzymes. Reaction - similarity - based splitting, based on SMILES sequence alignment, avoids the repetition of reactions in the training and test sets, thus effectively evaluating the model's prediction ability for new reactions.
In addition, ReactZyme introduced a negative - sample generation algorithm based on enzyme/reaction similarity. This method screens enzymes with highly similar structures or reactions but no actual catalytic relationships as negative samples, significantly enhancing the model's discriminative ability and enabling it to more accurately distinguish between real and false enzyme - reaction relationships.
Multi - Perspective Reaction Representation and the Empowerment of Uni - Mol
ReactZyme adopts a dual - channel representation method of 2D molecular graphs + 3D conformations, which can comprehensively describe the molecular characteristics during the reaction process from multiple perspectives.
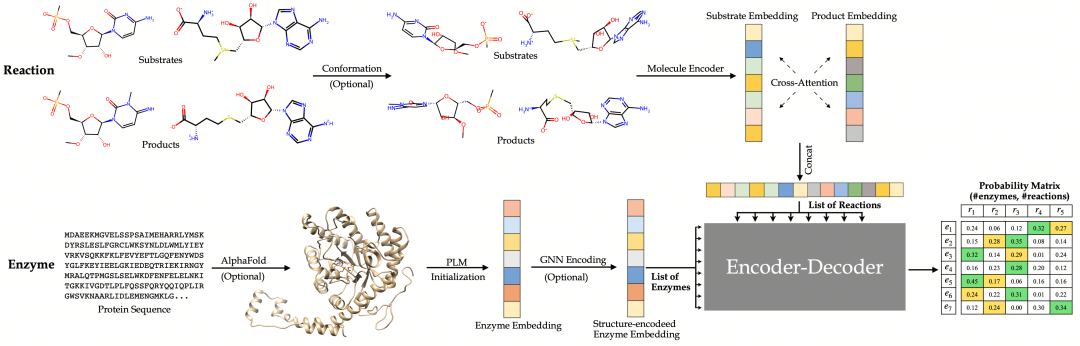
Figure 2 :First, calculate the conformational information of substrates and products according to the given reaction, and obtain the enzyme structure using AlphaFold. Then, transform the 2D molecular graph and its 3D geometric structure through a molecular encoder; a protein language model is used to initialize the enzyme features. Substrates and products are optimized and merged into a single reaction representation through the cross - attention mechanism, and the enzyme features are further optimized using an equivariant graph neural network (equivariant - GNN). Finally, the embedding representations of the enzyme and the reaction are input into an encoder - decoder model to establish pairwise relationships and calculate the probability matrix between the enzyme and the reaction for retrieval.
SMILES (Simplified Molecular Input Line Entry System) and molecular graphs play important roles. SMILES is a linear symbolic representation method for chemical molecular structures. It can concisely encode the topological structures of substrates and products, clearly showing the connection patterns of atoms and the changes of chemical bonds in molecules, providing intuitive information for studying the reaction process. Molecular graphs regard atoms as nodes and chemical bonds as edges. Through this graphical representation, the structures of molecules and the interactions between atoms can be more intuitively presented, which helps to capture the relationship between molecular structures and reactions.
Uni - Mol - 3D conformational modeling is one of the key technologies of ReactZyme. Through molecular force field optimization, Uni - Mol - 3D can generate the 3D coordinates of molecules, accurately capturing the spatial chemical environment of molecules. For example, it can accurately obtain important 3D structural information such as bond angles and steric effects. Experiments show that after introducing the 3D conformational features of Uni - Mol, the Top - 1 accuracy of the model is increased by 5.2%. This improvement fully demonstrates the effectiveness of Uni - Mol - 3D in capturing molecular spatial features and provides strong support for more accurately predicting enzyme - reaction relationships.
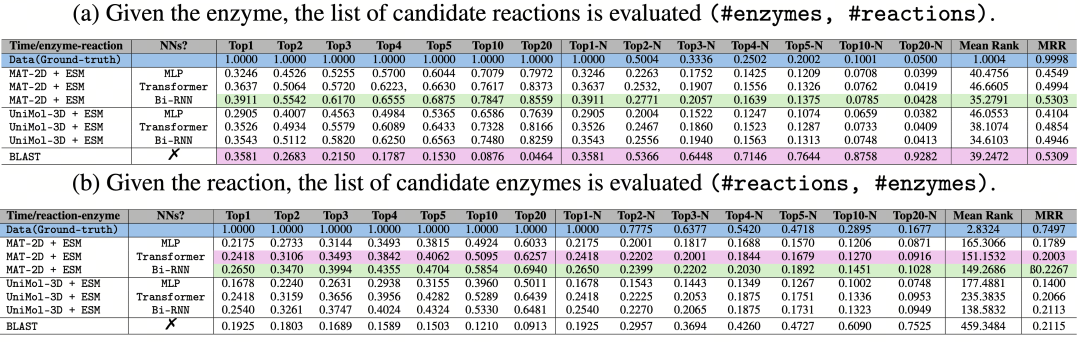
Table 2 : Average results of baseline models based on time - based division. (a) Given an enzyme, evaluate the list of candidate reactions; (b) Given a reaction, evaluate the list of candidate enzymes. The optimal results are highlighted in green, orange, and purple respectively.
.png)
.png)
Protein Language Models and Equivariant Graph Neural Networks
In the characterization of enzymes, ReactZyme integrates two innovative technologies, which greatly enhance the ability to understand and predict enzymes.
The structure-aware protein model SaProt is one of the important technologies. SaProt aligns the structural information of proteins through FoldSeek and can deeply analyze the sequence features of proteins, generating 1280-dimensional residue features. These features contain rich information about the structure and function of proteins, which helps to more accurately describe the characteristics of enzymes. Compared with traditional protein language models, SaProt can better capture the relationship between protein structure and function, providing a more accurate feature representation for subsequent enzyme-reaction prediction.
The equivariant graph neural network (SE(3)-GNN) is also an important part of ReactZyme. This network uses the three-dimensional structure of enzymes predicted by AlphaFold and realizes the description of rotational and translational invariance through frame averaging processing. This learning method enables the model to stably recognize the structural features of enzymes under different spatial transformations, without being affected by the rotation and translation of enzyme molecules in space. As shown in Table 3, the model combining SaProt and GNN achieves a Mean Reciprocal Rank (MRR) of 0.88 in the splitting of enzyme similarity, which is significantly better than the traditional ESM model. This result fully demonstrates the powerful advantages of this fusion technology in enzyme function prediction and can more accurately evaluate the matching degree between enzymes and reactions.
Table 3: Average results of the baseline model based on the division of enzyme similarity. The optimal results for each item are highlighted in green, orange, and purple.

.png)
Research Results: Performance Breakthroughs Beyond Classical Methods
Comprehensive Leading Performance in Benchmark Tests
ReactZyme has demonstrated outstanding performance in benchmark tests, comprehensively surpassing traditional methods. In the time-split test (see Table 2 above), the Uni-Mol-3D + ESM model has performed remarkably, refreshing the record with a Top-1 accuracy of 35.88% and an average ranking of 32.7. This achievement indicates that the model has high accuracy and reliability when predicting the relationship between known reactions and enzymes. When facing the prediction of new reactions (reaction similarity splitting), the ReactZyme model still maintains a Top-1 accuracy of 9.41%. In contrast, the traditional BLAST method's prediction results are nearly random (Top-1≈0%) in this situation. This significant gap fully demonstrates the advantage of ReactZyme in dealing with the prediction of new reactions, proving that its model has stronger generalization ability and adaptability.
Highlighting the Value of Three-Dimensional Structure
The comparative experiment (Table 4) further validates the significant value of the three-dimensional structure in enzyme function prediction. The model using the three-dimensional conformation has an AUROC (Area Under the Receiver Operating Characteristic Curve) of 0.8747, which is a 2.1% improvement compared to the two-dimensional model. This improvement shows that the three-dimensional conformation information can help the model more accurately identify the interaction between enzymes and substrates. In the prediction of the dynamic binding of enzymes and substrates, Uni-Mol-3D plays a crucial role. By capturing the geometric changes in the transition state, Uni-Mol-3D has increased the Mean Reciprocal Rank (MRR) by 19.3% (Table 5). This result fully illustrates the importance of the three-dimensional structure in understanding the dynamic binding process of enzymes and substrates, and also reflects the success of ReactZyme in using three-dimensional structure information to improve prediction performance.
Table 4: Average accuracy and the area under the receiver operating characteristic curve (AUROC) of the baseline model for enzyme-reaction prediction. The optimal results for each item are highlighted in green, orange, and purple.
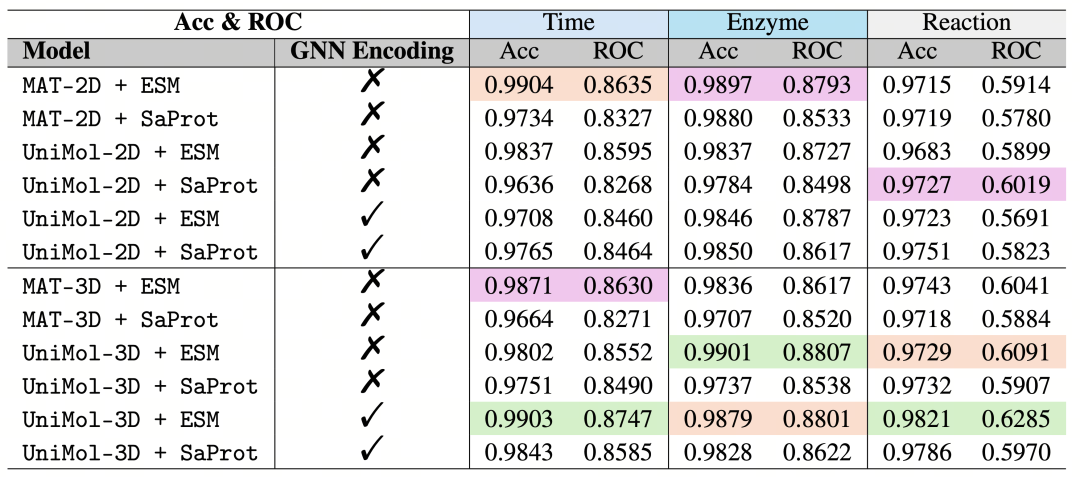
Table 5: Comparison between the neural network based on time division and the BLAST method. (a) Evaluating reactions given enzymes; (b) Evaluating enzymes given reactions.
Potential for Industrial Applications
ReactZyme has shown great potential in industrial applications and has successfully achieved two important scenarios.
In the mining of enzymes for new reactions, ReactZyme has recommended candidate enzymes for 386 unseen reactions. This achievement provides valuable references for fields such as biosynthesis and drug research and development, helping researchers discover new enzyme catalysts and promoting innovative development in related fields.
In the reannotation of enzyme functions, ReactZyme has identified potential catalytic functions among 12,277 unannotated enzymes (see Figure 2 above). This function can help researchers have a deeper understanding of the roles of these unannotated enzymes, improve the enzyme function database, and provide important basic data for subsequent enzyme research and applications.
Conclusion
In summary, the introduction of ReactZyme not only provides a larger-scale and finer-grained benchmark of enzyme-reaction data for academic research but also opens up a new path for the practical applications of biosynthesis and enzyme engineering. With the more comprehensive protein-molecule structure characterization ability and a flexible and efficient retrieval-based prediction framework, we are expected to have a deeper analysis of the catalytic mechanism of enzymes, screen out potential "star enzyme species", and contribute to key breakthroughs in the fields of drug research and development and green chemistry. Among them, Uni-Mol, as the core foundation for three-dimensional molecular modeling, not only significantly improves the model's ability to understand the complex structural relationships of enzyme-substrate complexes but also provides strong support for accurately capturing key catalytic features. Looking to the future, if the Uni-Mol framework can be further deeply integrated with multiple protein language models, it is expected to achieve a leap in the accuracy and efficiency of enzyme-reaction prediction, opening up broader imaginative space for the exploration and directional design of functional enzymes.