What Can Uni-Mol Do too? | Multi-objective optimization unlocks a new evolutionary approach to chemical product design
On July 15, 2024, Bilal Aslan from the University of Cape Town, Flavio Correa da Silva from the University of São Paulo, and Geoff Nitschke from the University of Cape Town collaborated to present their research titled “Multi-Objective Evolution for Chemical Product Design” at the Genetic and Evolutionary Computation Conference (GECCO). This study introduced a chemical product design method based on multi-objective evolutionary optimization. By innovatively integrating deep learning with evolutionary algorithms, the approach optimizes molecular properties and utilizes the Uni-Mol model to evaluate molecular toxicity, providing a novel solution for the design and optimization of chemical products.
Research Background
In the field of chemical product design, optimizing target properties within molecular structures has always been a highly challenging task. Traditional optimization methods rely primarily on laboratory experiments, which are not only time-consuming and costly but also struggle to identify ideal molecular candidates within the vast design space of chemical molecules. With the advancement of computational technologies, methods based on deep learning and multi-objective evolutionary optimization have gradually emerged in chemical product design, enabling the rapid generation and screening of molecules that meet specific property requirements.
However, these computational methods face challenges in comparing technical performance, as they must consider both quantitative evaluation and the diversity and innovation of generated molecular candidates. To address this, this study proposes a multi-objective evolutionary optimization framework that combines quantitative and qualitative evaluations. This approach aims to provide a more comprehensive comparison of different optimization techniques, facilitating more efficient molecular property optimization in chemical product design while ensuring that the generated molecules not only meet the required criteria but also exhibit innovation.
Uni-Mol Assists in Toxicity Assessment for Chemical Product Design
In this study, Uni-Mol was employed to evaluate the aquatic toxicity of molecules in chemical product design. Uni-Mol is a universal 3D molecular representation learning framework, pretrained on the 3D structures of over 2.1 billion molecules. The research team utilized an RDKit-based pretrained 3D structural model and fine-tuned the Uni-Mol model using data extracted from the publicly available PubChem database, creating a model specifically designed for molecular toxicity assessment.
During the molecule generation process, a multi-objective optimization method based on evolutionary algorithms was adopted. Initially, a set of seed molecules was selected from the PubChem database with a minimum similarity threshold of 80%. Uni-Mol was then used to evaluate molecular toxicity, excluding molecules with potential aquatic toxicity. The optimization process iteratively generated new molecules, selecting parent molecules based on the similarity threshold and generating offspring molecules according to hyperparameters (β and λ). The newly generated molecules were screened based on target properties, including minimizing molecular weight and complexity, maximizing XLogP, and ensuring non-toxicity. The optimization continued for multiple generations until reaching stability criteria or a runtime limit, ensuring that the resulting molecules met both functional and safety standards.
This innovative application demonstrates the robust capability of Uni-Mol in molecular toxicity prediction. Figure 2 illustrates box plots of molecular properties such as complexity, molecular weight, XLogP, and reference similarity, all of which are linked to the optimization process (including toxicity assessment). The data indicate that these properties fluctuated toward target values during the optimization process, indirectly supporting Uni-Mol's role in molecular screening. This study highlights Uni-Mol's significant contribution to the safe design of chemical products.
Introduction to Multi-Objective Evolutionary Optimization (MOEO) Methods
The implementation of multi-objective evolutionary optimization (MOEO) in chemical product design involves the following steps:
Optimization Objectives:
The optimization aims to minimize molecular weight and complexity, ensure the optimal XLogP value, and eliminate molecules with aquatic toxicity.Seed Molecule Selection:
The process begins by selecting seed molecules with a similarity of at least 80% to reference molecules. The search space is defined around these seed molecules to ensure that candidate molecules retain characteristics similar to the seeds.Toxicity Assessment:
A Uni-Mol-trained model is used to evaluate the aquatic toxicity of molecules, filtering out those with potential toxicity to ensure safety.Multi-Objective Optimization Process:
Multi-objective optimization strategies, such as MO-CMA-ES, are applied to generate new candidate molecules based on the similarity to seed molecules. Parameters such as similarity thresholds, parent selection, and offspring selection are controlled to iteratively optimize the candidate set of molecules, ensuring their performance aligns with the defined objectives.
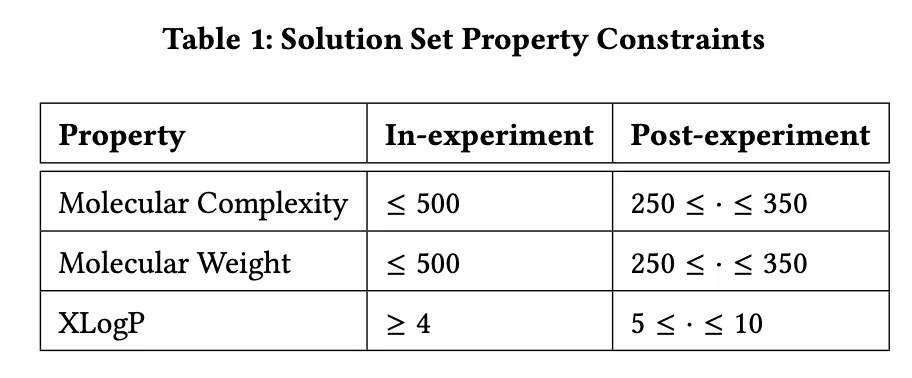
Table 1: Attribute Constraints for Chemical Product Design
Table 1 presents the attribute constraints used in the experiment for chemical product design. The table is divided into two parts: In-experiment Constraints and Post-experiment Constraints.
Molecular Complexity:
- In-experiment: Molecular complexity is constrained to be ≤ 500.
- Post-experiment: Optimized molecular complexity must range between 250 and 350.
Molecular Weight:
- In-experiment: Molecular weight is limited to ≤ 500.
- Post-experiment: Optimized molecular weight must range between 250 and 350.
XLogP (Lipophilicity Indicator):
- In-experiment: XLogP must be ≥ 4.
- Post-experiment: Optimized XLogP values must range between 5 and 10.
These constraints are designed to ensure that the molecules generated during the optimization process exhibit both good performance and practical applicability. The in-experiment constraints are relatively lenient, allowing for broader exploration in the early stages. In contrast, the post-experiment constraints are stricter, ensuring that the final molecules meet specific standards for chemical product design.
Results and Discussion
This section focuses on analyzing the effectiveness of applying the Multi-Objective Evolutionary Optimization (MOEO) method in chemical product design. The research team conducted 50 generations of evolutionary runs with 20 independent experiments to evaluate each method's performance in escaping local optima and promoting exploration. The results are summarized as follows:
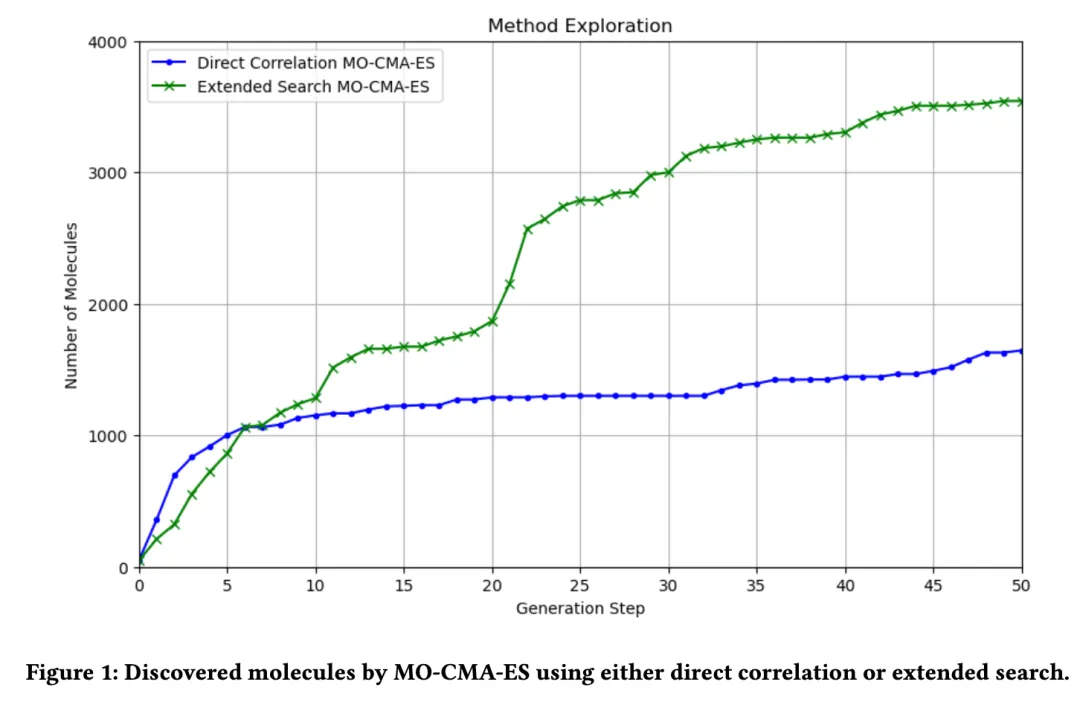
1. Diversity:
Figure 1 illustrates the variation in the number of molecules generated during the evolutionary process using the MO-CMA-ES algorithm under two approaches: directly related search and extended search.
Directly Related Search:
This method generates fewer molecules, primarily searching around the initial seed molecules within a limited search space. As a result, molecular diversity is relatively low.Extended Search:
Extended search allows the discovery of a larger number of molecular candidates, with the number of generated molecules gradually increasing throughout the evolutionary process. This demonstrates the method's advantage in expanding the search space and enhancing molecular diversity.
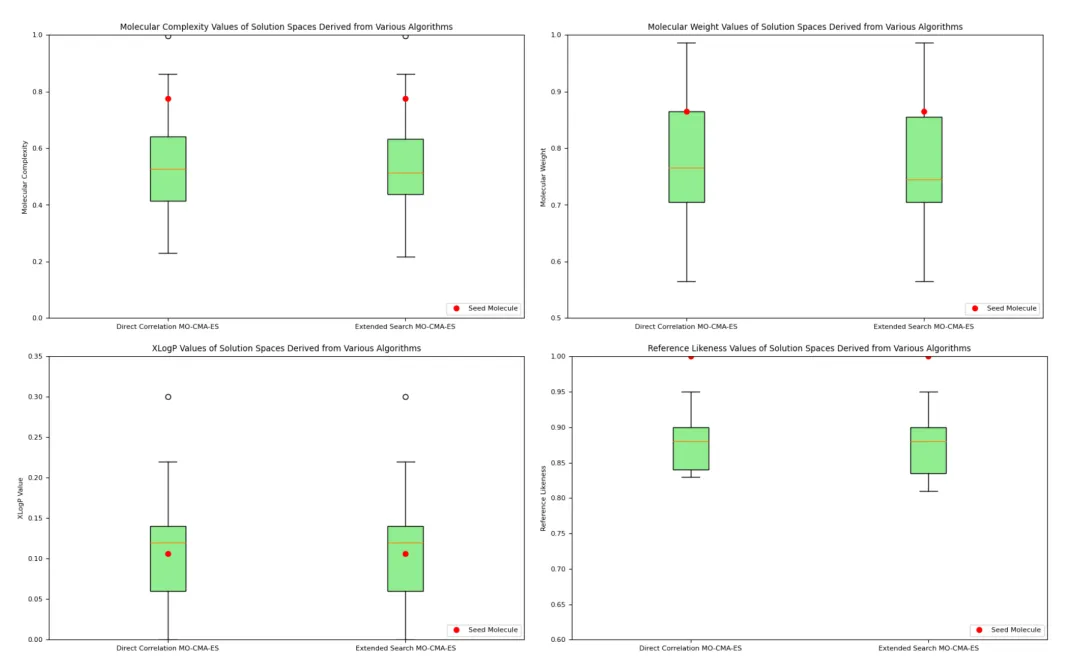
2. Quality of Optimized Solution Sets:
Figure 2 shows that after optimization using MO-CMA-ES, the properties of different molecules were well-controlled. While some properties (e.g., XLogP) exhibited significant fluctuations, the overall performance of the optimized molecules showed minimal variation within the target range, highlighting the effectiveness of the optimization method in maintaining both molecular characteristics and diversity.
Top Left: Molecular Complexity
This plot shows the distribution of optimized molecular complexity. The low height of the box plot indicates that the molecular complexity of the optimized molecules was concentrated within a narrow range, demonstrating the effectiveness of multi-objective optimization in reducing molecular complexity.Top Right: Molecular Weight
The distribution of molecular weight was similarly optimized to a narrow range, indicating that the optimization method effectively controlled molecular weight to meet the expected standards.Bottom Left: XLogP (Lipophilicity)
The XLogP values showed relatively large fluctuations but remained within the target range. The wider distribution indicates that more molecular candidates were explored during the optimization process.Bottom Right: Reference Similarity
Reference similarity measures the similarity between optimized molecules and seed molecules. The results indicate high similarity for most generated molecules, showing that the key characteristics of the reference molecules were retained.
These results highlight the MOEO method's ability to balance the trade-off between molecular diversity and optimized property control, effectively supporting the chemical product design process.
Conclusions
This work compares two heuristic search methods for multi-objective optimization (MOO) applied to chemical product design: directly related search and extended search. The experimental results demonstrate that extended search improves the diversity of solution sets without compromising solution quality. The primary contribution of this study lies in the development of appropriate evaluation metrics to compare different optimization strategies, particularly by assessing solution set diversity and quality.
Uni-Mol played a critical role in the study, especially in molecular toxicity prediction. By leveraging the trained Uni-Mol model, the research effectively filtered out molecules with aquatic toxicity, ensuring that the optimization results not only achieved performance breakthroughs but also met safety standards.
The results further revealed that the chemical design space is unevenly distributed in terms of molecular similarity and corresponding observable property values. This unevenness, particularly in multi-objective optimization scenarios, often leads to entrapment in local optima. Future work will focus on enhancing dynamic parameter optimization, fine-tuning parameter interactions (such as 𝛽 and 𝜆), and expanding the dataset using generative adversarial networks (GANs) and evolutionary transfer learning. These approaches aim to avoid local optima and generate more diverse solutions.