What Can Uni-Mol Do too? | Breaking the Limits of Few-shot Molecular Property Prediction
On August 19, 2024, Linlin Hou, Hongxin Xiang, and Xiangxiang Zeng from Hunan University, in collaboration with Li Zeng from the Shanghai Institute of Materia Medica, published a research article titled "Attribute-guided Prototype Network for Few-shot Molecular Property Prediction" in Briefings in Bioinformatics. This study introduced an Attribute-guided Prototype Network (APN), which innovatively combines high-level molecular fingerprints with deep learning algorithms, significantly improving the accuracy of molecular property prediction in limited-sample scenarios. This breakthrough opens a new direction for few-shot learning in drug development.
Research Background
Molecular property prediction (MPP) is a critical step in the drug discovery process. However, traditional deep learning methods often rely on large amounts of labeled data, making their application in few-shot scenarios highly challenging. Recently, a research team proposed an innovative Attribute-guided Prototype Network (APN), which overcomes the limitations of few-shot learning by extracting and leveraging high-dimensional molecular fingerprint attributes. Compared to existing methods, APN not only efficiently captures both local and global information within molecular structures but also demonstrates stronger generalization capabilities. This study offers new possibilities for molecular property prediction under limited data conditions, significantly accelerating virtual screening and drug optimization processes.
Uni-Mol Assists in Few-shot Molecular Property Prediction
In this study, Uni-Mol, a self-supervised learning model, was employed to extract deep attributes from molecular structures. These deep attributes, utilized in training neural networks, enabled the model to capture high-level molecular features, providing strong support for few-shot molecular property prediction. Specifically, Uni-Mol generated multiple molecular conformations (using 10 conformations in unimol_10conf) and, combined with Graph Attention Networks (GAT) and other molecular fingerprint data, optimized the local and global representations of molecules.
Experimental results demonstrated that Uni-Mol achieved outstanding performance on several datasets, including Tox21, significantly enhancing the prediction accuracy in few-shot learning scenarios.
Methods
Attribute-guided Prototype Network (APN)
Attribute-guided Prototype Network (APN) Architecture
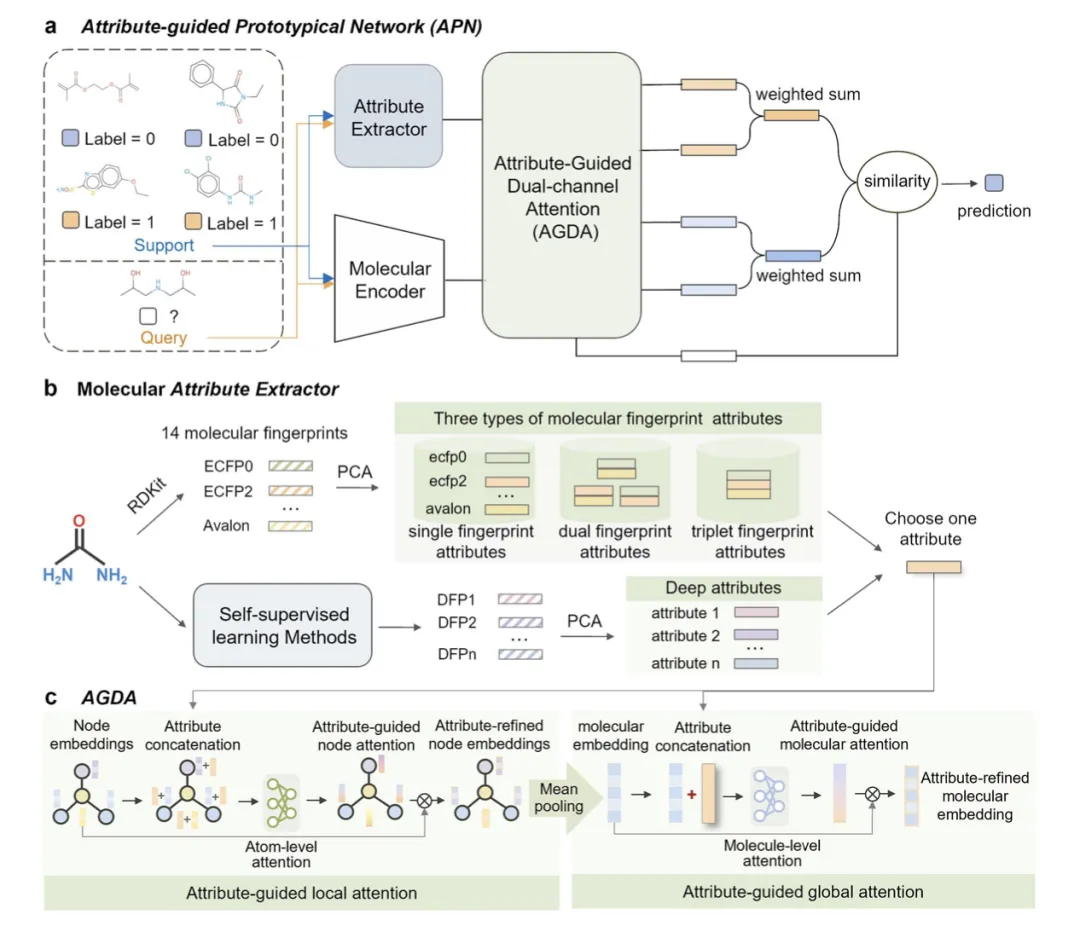
Figure 2 introduces the overall architecture of the Attribute-guided Prototype Network (APN), consisting of a molecular attribute extractor and an Attribute-Guided Dual-channel Attention (AGDA) module.
Figure 2(a) - Overall APN Architecture:
This panel illustrates the APN architecture using a 2-class, 2-shot task from the Tox21 dataset as an example. APN is optimized over a series of training tasks. In each task, a support set is used to derive prototypes for each class, while a query set is used to optimize the parameters of the molecular encoder and the AGDA module. The query molecule, represented as ( z' ), is generated via the molecular encoder and AGDA module, and its similarity to the prototypes is computed for final prediction.Figure 2(b) - Molecular Attribute Extractor:
This panel shows how single-fingerprint, double-fingerprint, and triple-fingerprint attributes are extracted from 14 types of molecular fingerprints. These attributes are further enriched using self-supervised learning methods to derive deep attributes, which are used to optimize molecular representations.Figure 2(c) - Detailed Structure of the AGDA Module:
The AGDA module employs both local and global attention mechanisms to optimize atomic-level and molecular-level representations.- The local attention module guides the model to focus on important local information.
- The global attention module helps the model capture overall molecular details.
Together, these mechanisms refine the molecular representation, improving its discriminative power for classification tasks.
This architecture effectively combines local and global molecular attributes to enhance performance in few-shot molecular property prediction.
Results and Discussion
Experimental Validation and Performance Comparison of APN with Baseline Models
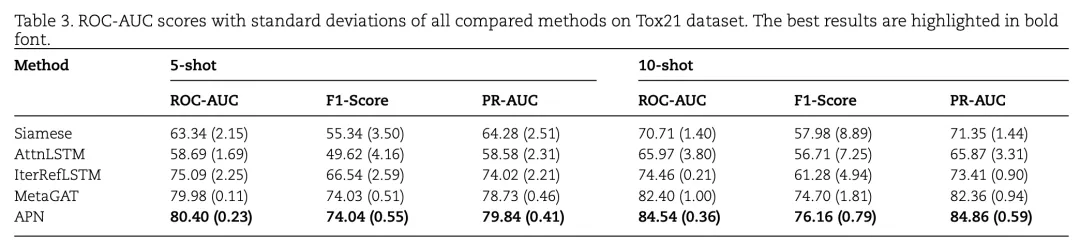
Table 3 compares the performance of the APN model on the Tox21 dataset with several baseline models, including Siamese networks, Attention LSTM, Iterative LSTM, and MetaGAT, for 5-shot and 10-shot tasks. The results show:
- 5-shot Task: APN achieves a ROC-AUC score of 80.40%, outperforming all other models.
- 10-shot Task: APN reaches a ROC-AUC score of 84.54%, again surpassing all baseline models.
- APN also leads in F1-Score and PR-AUC scores across multiple tasks, particularly excelling in the 10-shot task.
Analysis of Single-Fingerprint Attributes in APN
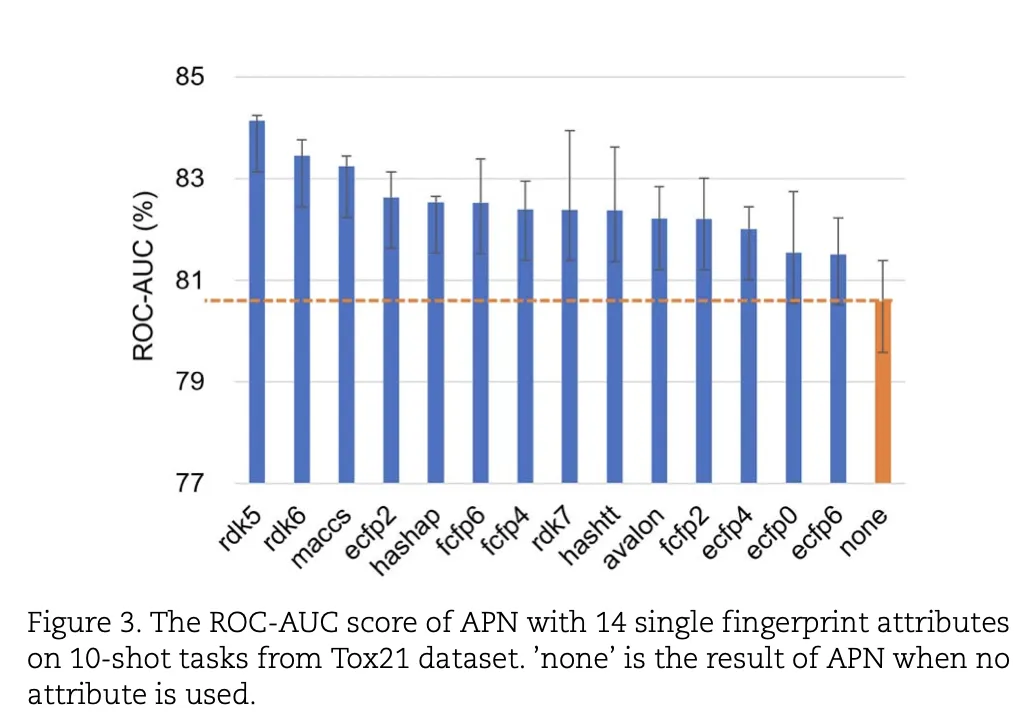
Figure 3 presents the ROC-AUC scores of APN when using 14 different single-fingerprint attributes for the Tox21 dataset in the 10-shot task:
- Each bar represents the ROC-AUC score for a single fingerprint attribute.
- Compared to the baseline without any attributes (“none”), using single-fingerprint attributes significantly improves APN's performance, with a maximum gain of 3.55%.
- Fingerprint attributes such as RDK5, RDK6, and HashAP, which are path-based, perform exceptionally well, significantly enhancing prediction accuracy.
Deep Attributes in APN
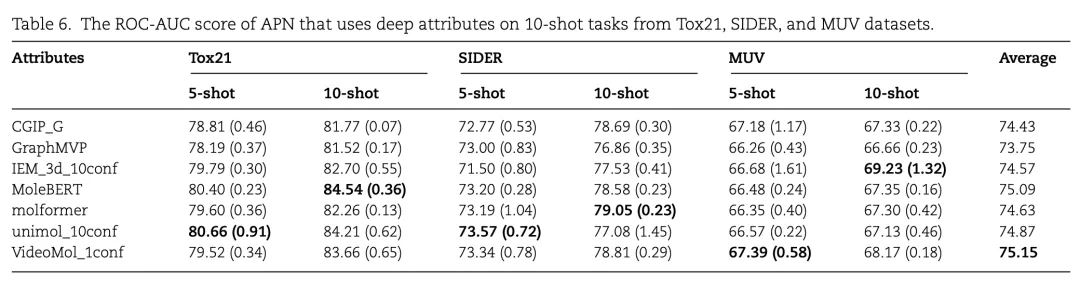
Table 6 demonstrates the performance of APN on the Tox21, SIDER, and MUV datasets using different deep attributes generated via self-supervised learning. Key findings:
Tox21 Dataset:
- Using the unimol_10conf deep attribute, APN achieves a ROC-AUC of 84.21%, F1-Score of 73.57%, and PR-AUC of 77.08%, showcasing strong predictive capability.
SIDER Dataset:
- The best-performing deep attribute is CGIP_G, with a ROC-AUC of 78.69%, while other attributes yield similar results.
MUV Dataset:
- The IEM_3d_10conf deep attribute achieves a PR-AUC of 69.23%, highlighting its potential for virtual screening tasks.
These results confirm that deep attributes effectively capture complex relationships between molecular structures and properties. In some cases, they even outperform traditional molecular fingerprint attributes, demonstrating APN’s strong generalization capability in few-shot molecular property prediction tasks.
Ablation Study on APN Modules
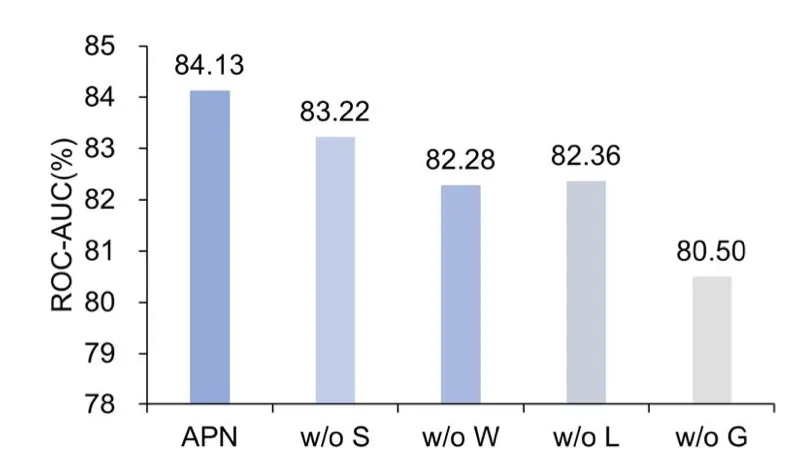
Figure 7 showcases the ablation study results on the Tox21 dataset, validating the effectiveness of different APN modules:
Global Attention Module:
- Removing the attribute-guided global attention module (w/o G) significantly reduces performance, emphasizing its critical role in capturing relevant information for few-shot tasks.
Local Attention Module:
- Removing the attribute-guided local attention module (w/o L) also decreases performance, though to a lesser extent than the global module, demonstrating its contribution to improved results.
Similarity and Weighted Prototypes:
- Variants without dot-product similarity (w/o S) and weighted prototypes (w/o W) underperform compared to the complete APN model, underscoring the importance of integrating these components.
These experiments reveal the synergistic effects of the global and local attention mechanisms and highlight the contribution of each module to overall performance improvement.
Performance Enhancement with Three-Fingerprint Combinations
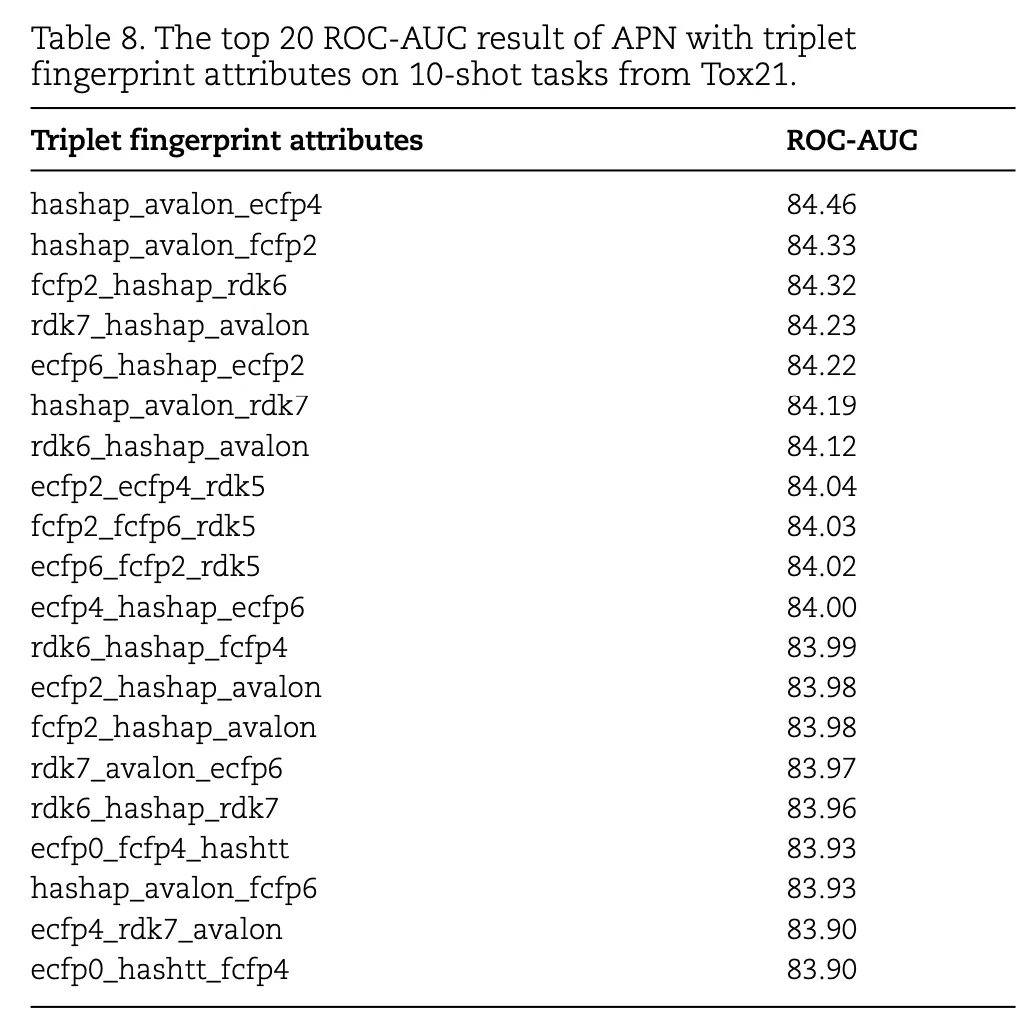
Table 8 lists the top 20 three-fingerprint combinations on the Tox21 dataset and their ROC-AUC scores:
- The best combination, hashap_avalon_ecfp4, achieves a ROC-AUC of 84.46%, outperforming both single and double-fingerprint attributes.
- Other effective combinations include hashap_avalon_fcfp2 and fcfp2_hashap_rdk6, with scores of 84.33% and 84.32%, respectively, demonstrating the potential of combining multiple fingerprint attributes.
These results validate that using three-fingerprint combinations significantly enhances prediction performance. Combinations involving attributes like HashAP and Avalon are particularly effective, showcasing the synergistic benefits of integrating multiple molecular fingerprints for few-shot molecular property prediction tasks.
Conclusions
In this study, the authors proposed the Attribute-guided Prototype Network (APN), which integrates high-level molecular attributes with deep attributes generated through self-supervised learning to address the challenges of few-shot molecular property prediction (FS-MPP). Uni-Mol played a critical role in this model. As a self-supervised learning framework, Uni-Mol generates deep attributes that effectively capture the complex relationships between molecular structures and properties.
By combining deep attributes generated by Uni-Mol (e.g., unimol_10conf) with traditional molecular fingerprint attributes, APN demonstrated outstanding performance across multiple datasets, including Tox21, SIDER, and MUV. The model significantly improved prediction accuracy, particularly in few-shot scenarios. Experimental results also revealed that APN, with its integration of local and global attention mechanisms, possesses strong generalization capabilities, making it highly effective for various molecular property prediction tasks.
Future research could explore additional combinations of deep attributes and molecular fingerprints to further enhance the predictive power and applicability of FS-MPP, broadening its value in real-world applications.