What Can Uni-Mol Do too? | Unveiling DeepGlycanSite: Precise Prediction of Carbohydrate Binding Sites
On June 17, 2024, researchers Xi Cheng and Liuqing Wen from the Shanghai Institute of Materia Medica, Chinese Academy of Sciences, in collaboration with Dingyan Wang from Lingang Laboratory, published a study titled "Highly accurate carbohydrate-binding site prediction with DeepGlycanSite" in Nature Communications [1]. This research introduces DeepGlycanSite, a deep learning-based algorithm for predicting carbohydrate-binding sites on protein structures with high precision. By leveraging Uni-Mol, DeepGlycanSite achieves exceptional accuracy in identifying carbohydrate-binding sites, providing a powerful tool for studying carbohydrate-protein interactions.
1. Research Background
Carbohydrates are widely present on the surface of all living cells, interacting with various protein families, including lectins, antibodies, enzymes, and transport proteins. These interactions regulate diverse biological processes, such as immune responses, cell differentiation, and neural development. Understanding carbohydrate-protein interactions is therefore fundamental to developing carbohydrate-based therapeutics.
However, due to the structural diversity of carbohydrates, obtaining experimental data on carbohydrate-protein interactions remains challenging. Structural determination techniques commonly used in glycobiology, such as nuclear magnetic resonance (NMR) and X-ray crystallography, require pure, stable molecules of detectable sizes.
Small carbohydrates (e.g., glucose with a molecular weight under 200 Da) are difficult to detect in structural studies due to their low atom count. On the other hand, complex long-chain carbohydrates (e.g., oligosaccharides with molecular weights exceeding 1000 Da) often involve multiple conformational states, leading to heterogeneity. In both cases, carbohydrate-binding residues of proteins cannot be clearly defined from a structural perspective.
Thus, developing a reliable tool for predicting carbohydrate-binding sites is critical to advancing our understanding of carbohydrate-protein interactions.
2. Cutting-Edge Deep Learning Technology—DeepGlycanSite
DeepGlycanSite is an equivariant graph neural network (EGNN) model based on deep learning, combining geometric features of proteins with evolutionary information to outperform state-of-the-art methods. This model not only predicts binding sites for monosaccharides and disaccharides but also accurately identifies binding sites for oligosaccharides and nucleotides.
The success of this study lies in the precise understanding of carbohydrate chemical structures, a capability significantly enhanced by Uni-Mol, which plays a critical role in the model's performance.

3. How Does Uni-Mol Assist DeepGlycanSite?
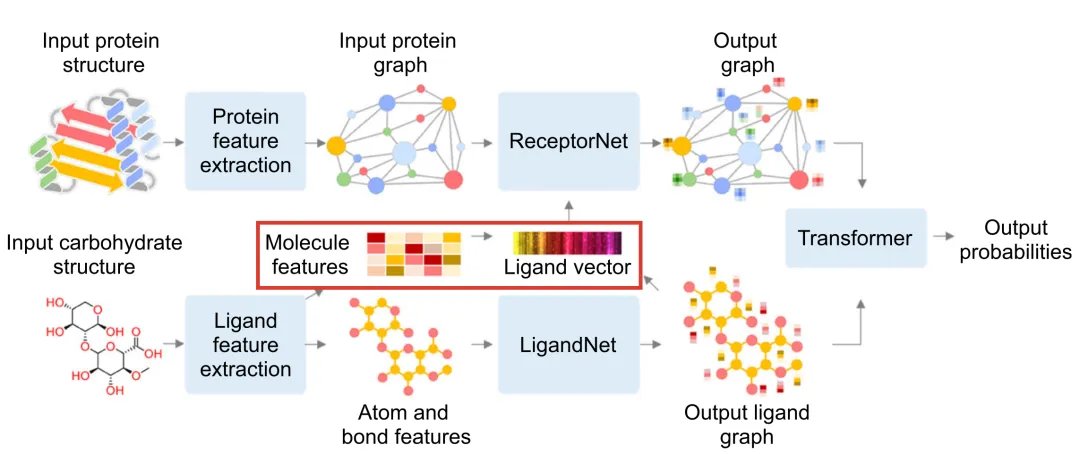
The performance of deep learning models heavily depends on the quality of feature extraction. In DeepGlycanSite, Uni-Mol is utilized to generate detailed chemical features of carbohydrates, enabling more accurate prediction of binding sites. The implementation is as follows:
3.1 Carbohydrate Processing
- SMILES Representation:
Rdkit is used to process the query carbohydrate and extract its SMILES representation. - Feature Generation:
Uni-Mol, integrated with Rdkit, converts the SMILES representation into molecular features.
3.2 Feature Extraction
Node Features:
Include detailed atomic properties:
- Atom symbol
- Degree
- Hybridization type
- Formal charge
- Number of radical electrons
- Aromaticity
- Total hydrogen count
- Chirality
Edge Features:
Capture bond-level information:
- Bond type
- Conjugation
- Ring membership
- Stereochemical configuration
Global Molecular Features:
Generate a 512-dimensional molecular feature vector encapsulating the overall chemical information of the carbohydrate.
3.3 Feature Integration
In the DeepGlycanSite+Ligand module:
- The ligand vector generated by Uni-Mol is fused with the protein graph’s node features.
- This integration is processed through an attention layer for feature alignment and updating.
- The combined features are then used to predict the binding probability of carbohydrates.
4. Experimental Validation and Results
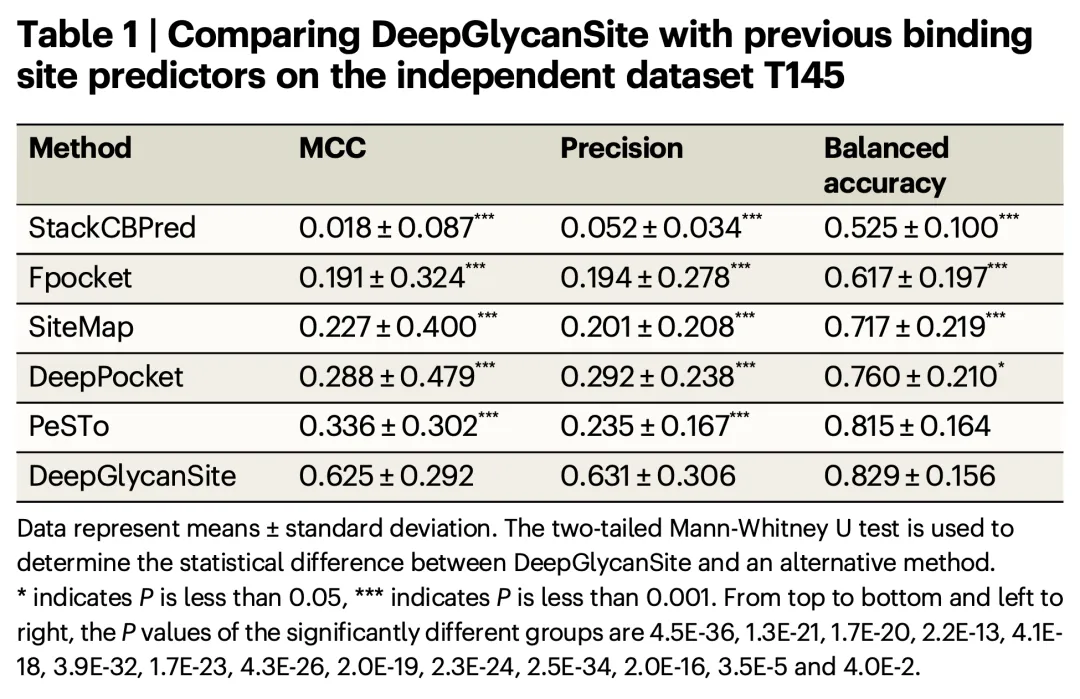
The study constructed a large dataset containing approximately 8,100 proteins and 1,700 carbohydrates and evaluated the performance of the DeepGlycanSite model on multiple independent test sets. The results demonstrated that DeepGlycanSite outperforms existing methods in detecting carbohydrate-binding sites.
- Key Metrics:
- Matthew’s Correlation Coefficient (MCC): 0.625 (average on independent test sets)
- Precision: 0.631
- Balanced Accuracy: 0.829
These metrics significantly exceed those of other comparison methods, highlighting the superior performance of DeepGlycanSite.
Conclusion
DeepGlycanSite is a highly efficient prediction tool that leverages Uni-Mol’s robust molecular representation capabilities to enhance the accuracy of carbohydrate-binding site predictions on proteins. By integrating sequence and structural information, DeepGlycanSite not only surpasses traditional methods in detecting monosaccharide or disaccharide binding sites but also excels in identifying multiple binding sites. This provides critical insights into carbohydrate-protein interactions.
Uni-Mol's ability to precisely capture chemical features and significantly improve predictive performance has established DeepGlycanSite as a powerful tool for addressing complex biological tasks. Its low dependence on protein structural accuracy enables analysis using predicted structures, supporting research into carbohydrate biological functions and drug development.
The study encourages researchers to explore Uni-Mol for various downstream applications in different domains. The team welcomes collaboration and discussion to unlock further possibilities!
Reference:
[1] He, X., Zhao, L., Tian, Y. et al. Highly accurate carbohydrate-binding site prediction with DeepGlycanSite. Nat Commun 15, 5163 (2024). https://doi.org/10.1038/s41467-024-49516-2
[2] Zhou G, Gao Z, Ding Q, Zheng H, Xu H, Wei Z, et al. Uni-Mol: A Universal 3D Molecular Representation Learning Framework. ChemRxiv. 2023; doi:10.26434/chemrxiv-2022-jjm0j-v4